Key Facts
- Ribosomal RNAs (rRNA) perform critical functions in the ribosome that allow protein synthesis to occur.
- The genes that encode rRNAs evolve (i.e. change sequence over time) in a very unique manner that makes them excellent “markers” to trace evolutionary history and powerful tools to identifying species from sequence data.
The Ribosome
The ribosome is a cellular machine found in all organisms. It serves to convert the instructions found in messenger RNA (mRNA, which itself is made from instructions in DNA) into the chains of amino-acids that make up proteins. That is, the ribosome is responsible for the synthesis of proteins.
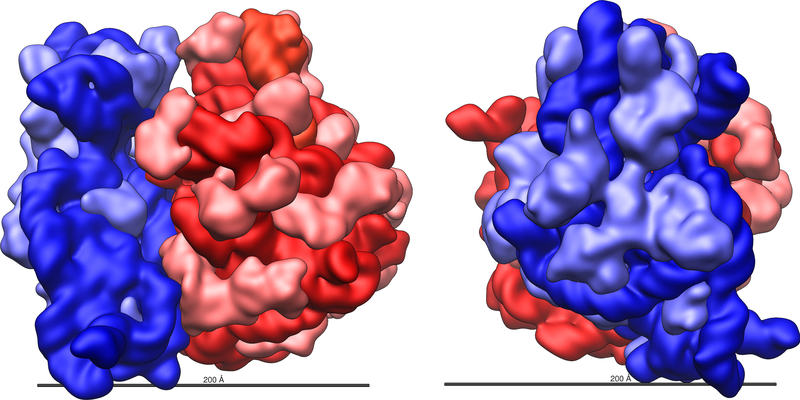
The ribosome itself is highly complex. It is made up of dozens of distinct proteins (the exact number varies a little bit between species) as well as a few specialized RNA molecules known as ribosomal RNA (rRNA). Note – these rRNAs do not carry instructions to make specific proteins like mRNAs. The ribosomal proteins are rRNAs are arranged into two distinct ribosomal pieces of different size, known generally as the large and small subunit of the ribosome.
(above text modified from the microBEnet factsheet on DNA-RNA-Protein)
The RNA World
The key catalytic activity of the ribosome – the creation of a chemical bond between two amino acids (known as a peptide bond) – comes from the RNA component of the ribosome. This, and other catalytic roles for RNA, were discovered relatively recently and were a bit of a surprise, since for many years it had been thought that all catalytic activities in cells were from proteins. The catalytic role of rRNA is a key piece of support for the “RNA world” hypothesis, which postulates that the first evolving chemical entities on Earth were RNA molecules. These RNAs served simultaneously as both genotype (i.e., genetic material) and phenotype (i.e., they had catalytic activities that allowed for some function). At some point in the evolution of life, some of these RNAs “discovered” DNA as a more stable storage medium for information, and proteins as a more versatile way to mediate enzymatic reactions. However, some catalytic functions of RNA (e.g., synthesis of proteins) are still present in modern organisms as a relic of earlier times.
Homology of the ribosome across life
Most of the individual components of the ribosome have equivalent counterparts across all forms of life. The matching components in different species (e.g., ribosomal proteins, or femur bones) are considered to be homologous – which means they are thought to have evolved from a common ancestor that also had that component (i.e. the ancestor also had a version of the same ribosomal protein).This is true for many of the proteins found in the ribosome and also for the ribosomal RNAs (for more detail see section below on Homology of ribosomal components. We can compare ribosomal components of different species, much as we can compare bones between different mammals, in essence “lining up” the sequences of the equivalent component to see where they differ.
Homology of ribosomal components
Homology refers to similarity of traits due to shared ancestry. The human hand is homologous to the cat paw and whale flipper. The three structures have evolved characteristics to different purposes, but share an underlying bone structure passed down from an ancestor common to all mammals.
Underlying the shared bone structures are shared genetic structures. Homologous genes encode homologous traits, from skeletons and organs down to the invisible components of cells. The ribosome is one of the oldest and most essential cellular players, and is homologous in all organisms. We infer homology of ribosomes across all life because they not only look and act alike, but also have remarkably similar gene sequences.
For example, the ribosome of the bacterium Escherichia coli includes a protein known as ribosomal protein L4 (“L” here stands for the fact that this protein is a component of the large subunit of the ribosome). In the genome of E. coli there is a region that includes the instructions for making this protein, known as the rpl4 gene. This gene is transcribed into mRNA, and the mRNA translated into the rpl4 protein. All organisms on the planet have a protein in their ribosomes comparable to L4 from E. coli. And, of course, all these organisms also have a gene in their genome that is equivalent to the rpl4 gene from E. coli (they could not have a L4 protein if they did not have a gene encoding it in their genome). In fact, it is not just that all organisms have a protein that is “like” the E. coli L4. All the L4-like proteins in all species are so similar to each other in their amino acid sequence, that it has been inferred that a common ancestor of all modern day organisms also possessed an L4-like protein.
The same pattern generally holds true for dozens of other ribosomal proteins, as well as for multiple ribosomal RNAs. We can line up the components of one species’ ribosome with the components of other species ribosomes much as we can compare bones between different mammals. From these comparisons it has been inferred that a common ancestor of all modern day organisms had a ribosome that was very similar to the ribosomes found across all forms of life today. The equivalent ribosomal components in different organisms (e.g., L4 in E. coli and humans) are considered to have evolved from a common ancestral component (i.e., L4 in a common ancestor of humans and E. coli). (Of course, at some point, long in the past, the ancestors of E. coli and humans split into different lineages – more on this “Tree of Life” in a bit).
The L4 proteins of different species are considered to be homologous.
Distinct rRNAs in different organisms
Bacteria and Archaea possess three distinct rRNAs, sometimes referred to as the 5S, 16S, and 23S forms. The “S” in this nomenclature refers to Svelberg units, a measure of an experimental technique called sedimentation (see next paragraph for more detail on this). The 16S rRNA is the sole rRNA in the small subunit of the ribosome and thus is sometimes referred to as the small subunit rRNA or ss-rRNA. The 5S and 23S are both components of the large subunit of the ribosome.
Sedimentation values
Sedimentation in the lab is in essence an accelerated form of the settling of particles that occurs in formation of sediment in lake and ocean floors. In the lab one can accelerate the process by very rapidly (10s of thousands of RPM) spinning samples in a centrifuge. To study the components of a cell such as the different parts of the ribosome, researchers break open cells and then spin the components in a tube inside a centrifuge. Different components of the cell settle in different regions of the “sedimentation gradient” in the tube (with the specific region given a numerical value – the higher the number the less easily something moves through the gradient). The exact region in which something settles is based on a combination of its size, shape and density. For most bacteria and archaea, the main forms of ribosomal RNA settle at the 5S, 16S, and 23S regions of a sedimentation gradient. For most eukaryotes, the main forms of ribosomal RNA settle at slightly different regions and thus have different numerical values (e.g., humans have 5S, 5.8S, 18S, and 28S and 40S. The 5.8S and 5S are homologous to the 5S of bacteria and archaea, the 18S is homologous to the 16S, and the 28S is homologous to the 23S.
Key features of rRNAs for phylogenetics
The function of rRNAs is very similar across all species. The core function of the ribosome is basically the same across different groups of organisms. However, this does not mean the rRNAs are identical between species. The actual sequence of the nucleotides in rRNAs (and in the rDNA genes) does vary between species. For our purposes there are three key features of the variation in rRNA sequence between species.
First, the rRNA molecules in the ribosome fold over into complex three dimensional shapes. The specific shape that they take is highly conserved between species. However, the linear sequence of nucleotides in each rRNA (also known as the primary sequence) can vary without affecting the function since different primary sequences can fold into in essence the same shape.
Second, when a single species splits into two distinct evolutionary lineages, differences can accumulate in the sequence of the rRNAs between the two lineages. Biologists call the process “sequence divergence.” The divergence of rRNA sequences generally occurs very slowly (they are among the most slowly evolving of genes, but changes still happen). In addition, the structure and function of the rRNAs generally stay the same between species (and changes in the actual structure are usually lethal).
Third, some regions of rRNAs evolve (i.e., diverge) slowly and others diverge rapidly. Some regions are basically the same across most or all taxa. This shared sequence has allowed researchers to use a specialized laboratory method known as polymerase chain reaction (PCR) to help read the sequences of rRNAs from different (and even unknown) species.
For background on protein synthesis and the ribosome, go here.
For information on the use of rRNA sequences in microbial ecology go here.
This document was produced by microBEnet. It was written by Jonathan Eisen and David Coil, and edited by Elizabeth Lester with feedback from Hal Levin.
RNA diagram from Wikipedia Commons.