While not about COVID19 directly, this paper “Initial Mapping of the New York City Wastewater Virome” has implications for the many many folks looking at SARS-CoV-2 in wastewater these days. This work is looking at all the viruses in the wastewater system and is one of relatively few metagenomic studies out there that focuses on …
This is a pre-print that just came out from several UC Davis colleagues. I actually think the title is a bit misleading because these recommendations are a lot broader than just human fecal metagenomes. Definitely worth a read! Abstract below: Background Shotgun metagenomes are often assembled prior to annotation of genes which biases the functional …
Job posting from here at UC Davis: The laboratories of Danielle Lemay (USDA Western Human Nutrition Research Center) and David Mills (UC Davis Food Science) seek a motivated postdoctoral scholar for a one year project. The goal of the project is to determine how dietary intake influences antimicrobial resistance in fecal metagenomes of healthy adults. The …
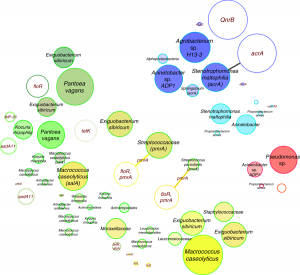
Antibiotic resistance has been assessed to rise to dangerously high levels in all parts of the world, and new resistance mechanisms are emerging and spreading globally. At the same time the number of people dying from antibiotic-resistant bacteria is increasing. The World Health Organization considers the spread of antibiotic resistance and appropriate countermeasures as one …
Just got pointed to a description of this 1-day meeting in New York on October 12, 2018… “It’s a Brave New World: Applications, Promise and Public Implications of Metagenomics in Urban Settings”. Sounds like a great lineup of speakers and certainly a fascinating topic! Meeting description below: This 1-day symposium brings together urban microbiome …
My collaborator, Marina LaForgia, and I are gearing up to start processing some plant rhizosphere samples for metagenomic sequencing. We collected more samples than we can afford to process (as is life) and are starting to make the hard decisions about which samples to move forward with. This brought us to the topic of whether …
So – just a quick post for now. More details to come. But I wanted to get a little bit about this out there. For a few years I have been teaching a mid-level course at UC Davis on “DNA sequencing based studies of microbial diversity’. The course has evolved over the years from a …
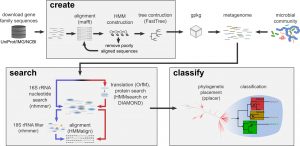
This could be of use to many people: Source: GraftM: a tool for scalable, phylogenetically informed classification of genes within metagenomes | Nucleic Acids Research | Oxford Academic Large-scale metagenomic datasets enable the recovery of hundreds of population genomes from environmental samples. However, these genomes do not typically represent the full diversity of complex microbial …
Genome-resolved metagenomics is the process of recovering de novo genomes belonging to individual organisms present in a community using DNA extracted from the whole community. These genomes are sometimes referred to as MAGs (metagenome-assembled genomes), and are recovered by associating assembled contigs together in “bins” in a process known as genome binning. As opposed to …
Of potential interest – new paper from the Banfield Lab – thanks to Elisabeth Bik for pointing me to this on Twitter. Preterm infants exhibit different microbiome colonization patterns relative to full-term infants, and it is speculated that the hospital room environment may contribute to infant microbiome development. Here, we present a genome-resolved metagenomic …