A new volume of Studies in Mycology was published recently and is dedicated to the diversity in the fungal genera Aspergillus, Penicillium and Talaromyces, all of which play a significant role indoors. The issue includes 6 papers related to our Indoor Mycota Barcode of Life (IM-BOL) project funded by the Alfred P. Sloan Foundation Program on the Microbiology of the Built Environment. Our goal is to bridge the knowledge gap between uncultured and cultured fungi in indoor environments by providing authoritative taxonomic and DNA barcode data to be used for metagenomic studies. House dust was collected from all over the world. Half the dust was used for isolations using a dilution-to-extinction (d2e) method, while the remaining dust was used for a 454-pyrosequencing analysis (published in Amend et al. 2010).
The first paper deals with the identification of the 2717 d2e isolates belonging to Aspergillus, Penicillium and Talaromyces. The usual view of indoor moulds recognizes about 40 common species of these genera, but we found 126 species of which 18 were described as new (Aspergillus 1160 isolates, 59 spp., 8 new; Penicillium 1459 isolates, 49 spp., 7 new; Talaromyces 98 isolates, 18 spp., 3 new). For each species, ITS barcodes and the secondary barcodes Calmodulin (Aspergillus) and β-tubulin (Penicillium & Talaromyces) were generated, adding to the reference sequences. Reference sequences obtained were not only from the commonly found indoor species, but also rare ones such as A. taichungensis, P. incoloratum and P. hispanicum for which only singleton sequences were available previously.
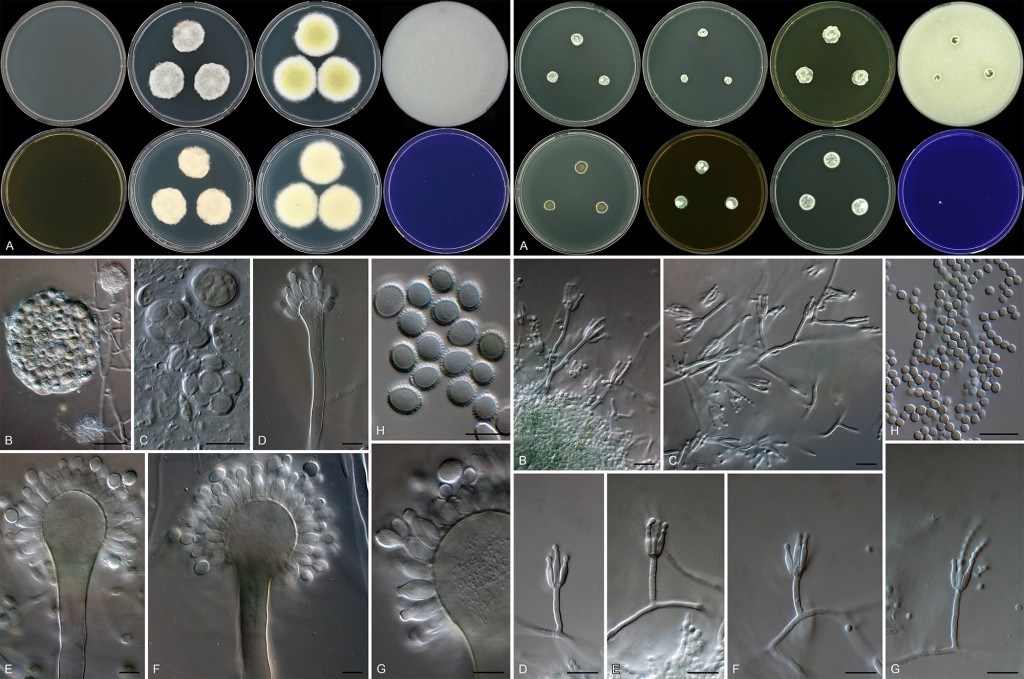
Among the interesting new species were an Aspergillus and Penicillium, which we found remarkable enough to name them after Alfred P. Sloan. Aspergillus has long been known to contain xerophilic species (previously also associated with the teleomorphic genus Eurotium), but the discovery of a species that is not able to grow on any of the media generally used for Aspergillus identifications was remarkable. The species only grew on low water activity media and then produced a eurotium-like sexual state and conidiophores with very large spiny conidia. This was our favorite Aspergillus isolated in the study and named it Aspergillus sloanii. The interesting Penicillium species was described as P. alfredii and was phylogenetically so distinct that it could not be placed into any of the 25 taxonomic sections of Penicillium. Unfortunately we could not introduce a new section for P. alfredii, because its phylogenetic position makes two other sections polyphyletic. This issue will be resolved in future, but additional data and sampling is needed to accomplish this.
Fig. 1. Aspergillus sloanii (left), an obligately xerophilic species only growing on low water activity media such as DG18. Penicillium alfredii (right) also displays slow growth on general media.
The remaining papers in the issue were purely taxonomic, but crucial for the framework of IM-BOL because they provide authoritative taxonomic information and reference sequences to be used for future identifications. In light of the move to one fungus = one name in the International Code of Nomenclature for algae, fungi and plants (ICN), the International Commission of Penicillium and Aspergillus (ICPA) decided to update the accepted species lists for these genera. From the start, we wanted these lists to be more than just nomenclature and thus we included MycoBank numbers, collection numbers to living reference cultures and GenBank accession numbers to reference sequences (ITS, β-tubulin, Calmodulin and RPB2). These lists were published in three papers dedicated to the taxonomy and nomenclature of each genus and can be downloaded here (Aspergillus, Penicillium, Talaromyces (the latter traditionally classified with Penicillium)). The lists are also hosted and curated online on the ICPA website, where updates will be posted. Most importantly, authoritative reference sequences are now available for the 339 accepted Aspergillus, 354 Penicillium and 89 Talaromyces species. The ITS data set was also included in the RefSeq initiative of NCBI.
The two remaining papers are on the important groups known as Aspergillus sect. Circumdati (common ochratoxin producers) and Penicillium sect. Aspergilloides (common indoor irritants such as P. glabrum and P. spinulosum). From d2e of house dust, a large proportion of strains belonged to these two groups, which are taxonomically very difficult. As such, we completed a monograph of Aspergillus sect. Circumdati, including descriptions and reference sequences for the 27 accepted species, 7 of them described as new, 3 of these from house dust. The Penicillium section Aspergilloides paper provided an extensive sequence data set for the 51 species, including 25 newly described species, 4 of them from house dust.
It is our continued goal to make identifications as easy as possible in these genera and we believe that Studies in Mycology 78 makes a big leap towards this goal. For IM-BOL, the next step is to use the reference sequences generated thus far and compare d2e communities with 454-pyrosequencing communities and determine just how far you can go with ITS sequences as identification tool within these genera. We will now start shifting focus onto xerophilic fungi, i.e. those that grow in arid, dry environments and can grow at low water activities, which our initial project revealed to be much more phylogenetically diverse than previously realized.
Contact information:
Cobus M. Visagie, Applied & Industrial Mycology, CBS-KNAW Fungal Biodiversity Centre, P.O. Box 85167, 3508 AD Utrecht, The Netherlands
Keith A. Seifert, Adjunct Professor, University of Ottawa, Dept of Biology, Gendron Hall, 30 Marie Curie, Ottawa ON K1N 6N5 Canada. Mailing address: Biodiversity, Eastern Cereal and Oilseed Research Centre, Agriculture & Agri-Food Canada, 960 Carling Ave., Ottawa Ont. K1A 0C6 Canada
Robert A. Samson, Head, Applied & Industrial Mycology, CBS-KNAW Fungal Biodiversity Centre, P.O. Box 85167, 3508 AD Utrecht, The Netherlands