Some antibiotics routinely used in the clinic are considered to have little side effects. Such is the case for cefprozil, a second generation cephalosporin. This relative safety may be true (to a certain degree) from the viewpoint of humans taking the antibiotic. However, it is not the case from our commensal microbes’ perspective. Our study, published last week in The ISME Journal, shows that cefprozil does indeed affect the microbiome in a manner that has the potential to both influence the activity of opportunistic pathogens and lead to the emergence of resistance genes. This may seem a big statement, but in fact lots of unexpected events can be observed in our microbiome if we look en profondeur.
The study design was straightforward. Eighteen healthy volunteers from Quebec City were given a seven day course of cefprozil, replicating what is prescribed by physicians for mild infections. Our first observations were coherent with a small footprint of cefprozil on the microbiome. Indeed, metagenomic sequencing showed that the most abundant members of the microbiome were not perceptively altered by the antibiotic. However, looking deeper, we observed that a significant number of low abundance microbes were negatively affected by intake of the antibiotics. But this is not what made our results so interesting (and a bit scary).
What really struck us as odd was that a third of the participants clustered together after the antibiotic treatment. What was so similar? They all had increased abundance of Enterobacter cloacae, an opportunistic pathogen known to favor the spread of antibiotic resistance genes. We thus asked the following question: are there properties of microbiomes that could explain this bloom of E. cloacae? We found that the answer was linked to enterotypes. Indeed, participants with E. cloacae had low diversity Bacteroides enterotype before exposure to the antibiotic while participants with high diversity microbiomes did not suffer proliferation of this opportunistic pathogen. Although further studies are needed to understand this phenomenon, we feel that it is reminiscent of what happens with Clostridium difficile after antibiotic treatment. Nonetheless, in our study, blooms of E. cloacae were not associated with clinical side effects in the healthy participants.
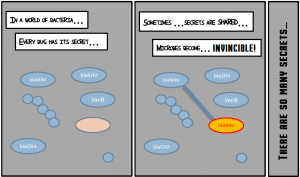
We also went further than just counting microbes by monitoring the gene content of microbiomes. The resistance gene content (the resistome) of individuals is even more variable than their species composition. Therefore, the resistome of people is so different that the impact of the antibiotic was specific to each person. After exposure to antibiotics, a different set of beta-lactamase resistance genes, unseen before treatment, appeared in each participant. We also observed important changes in resistance genes not related to beta-lactam resistance. Thus, even “low side effect” antibiotics can affect the resistome. Is it relevant clinically? Most of the time, probably not, but so many people take antibiotics that once in a while a new resistance gene can end up in a multiresistant pathogen.
So, where should we go from here? We call for studies of the impact of several antibiotics on the microbiome and the resistome. First, pharmaceutical companies should determine the impact on the microbiome of their antibiotics but also of their other drugs. Second, our results show that knowledge of the initial composition of the microbiome could help predict adverse effects. Third, we should consider our commensal flora as allies and seek an active partnership with our microbes. Acting along these lines are the next step towards a personalised medicine devised to make us live happier with our microbes.