It is well known that antibiotic resistance in bacteria happens much faster than we can possibly develop novel antibiotics. So what if instead of trying to reinvent the wheel, we just rearrange it? Well, researchers at the H. Lee Moffitt Cancer Center and Research Institute had a similar idea in regards to reducing antibiotic resistance that was recently published in PloS Computational Biology.
They use mathematical approaches to show that administering the same antibiotics in different orders results in a different final population. The idea being that you can purposefully select a certain kind of fitness with Drug A, and then use Drug B to attack the very mode which allowed the bacteria to survive Drug A. They argue that this approach steers evolution and prevents the emergence of antibiotic resistance.
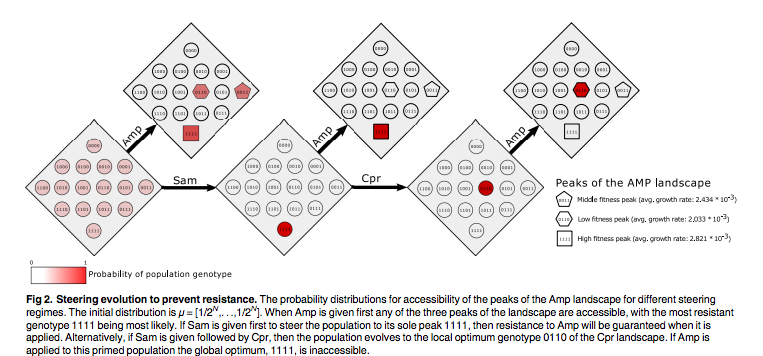
At first I was very impressed, but then I started to think about practical applications of this principal. Isn’t this just too little too late? The same pathogens that have sparked global concern about antibiotic resistance are the same ones that already have multiple forms of resistance. That is, it would be hard to find a sequence of antibiotics to use on a bug that already has several different adaptive mutations. How do you steer evolution in that kind of fitness landscape? This study just looked at B-lactam antibiotics, but numerous other classes of antibiotics are used excessively and at random all over the world.
What about clinical use of this principal? The authors theoretical analysis is too far removed from the practicality of this. If a patient’s infection is severe enough to need antibiotics, there may not be enough time to methodically steer the infection through adaptive mutations, assuming the evolutionary trajectory of it goes perfectly to plan. And what about spreading resistance indirectly during each antibiotic treatment? What about all of the other microbes in and on the patient — is this really the healthiest way to suggest treatment?