Guest post by Melina Bautista (@qmbautista) and Ameet J. Pinto (@watermicrobe).
The provision of safe drinking water relies primarily on disinfection as a cornerstone for pathogen control. Since its full-scale implementation in the early 20th century, it has become the standard final treatment step to inactivate microorganisms in most drinking water treatment plants around the world. We have known for a long time that safe drinking water is not sterile. Yet, the extent of microbial diversity in drinking water systems has only recently become apparent thanks to high-throughput DNA sequencing approaches. If we take a closer look at each individual drinking water system, there is heterogeneity from the source to the tap in each of them. Nevertheless, despite all this heterogeneity, we know that the concentrations of drinking water chemical parameters in most systems fall into specific ranges, some are health-based guideline values (i.e. related to potability) while others are related to palatability (e.g. taste, odour, colour). What about the microbial communities across these “chemically-similar” drinking water environments?
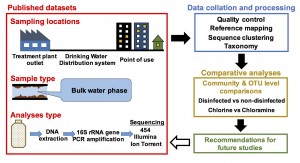
We have some knowledge on this subject, but the data that we have is from separate studies that used different approaches for sample collection, processing, analysis and reporting. To try to answer this question, we conducted a collective analysis or meta-analysis of microbial communities in full-scale drinking water systems. At the phylum and class levels, we found broad similarities between drinking water systems irrespective of type and presence/absence of disinfectant residual. But, as you start refining from phylum/class down to family or genus, microbial communities in these systems start looking different from each other. We also ran into the stumbling block that most meta-analysis studies of this type run into — data heterogeneity (sequencing platform, hypervariable region, etc.). So, we knew our findings would not be quantitative but mostly qualitative, to help identify indicative trends that could be important to follow-up on.
First, there were clear distinctions in terms of diversity in systems that maintain a disinfectant residual and those that don’t. This might not seem that surprising — sure, chemical stress in disinfected systems will impact diversity. But, disinfectant residual free systems tend to be more nutrient deprived (i.e. significantly lower growth substrates) than systems that are disinfected. So, this may be a nice follow-up area to dig deeper into in the context of drinking water systems — chemical vs. nutrient stress and microbial diversity, using controlled experiments. It was also interesting to see how abundant and frequently detected potentially predatory bacteria were across drinking water systems (e.g. Bdellovibrio). There is very little that has been done on understanding the dynamics of predatory bacteria in drinking water systems, which is somewhat surprising given that their likely technological application is quite clear (e.g. biological disinfectant or biofouling control in filtration systems). We also looked at potential opportunistic pathogens (OPPs) across datasets, but the big caveat is that partial 16S rRNA gene sequence (or in some cases even full 16S rRNA gene) is not sufficient to confidently identify OPPs. However, we felt it was important to dig deeper into this aspect even if it was solely for the purpose of generating hypotheses about the effect of the use and type of disinfectant residual on potential OPPs and we discuss the differences in the paper.
Data heterogeneity was a big limitation for our paper. On the chemistry side of things, most drinking water studies do a rigourous job of analyzing multiple water quality parameters and use protocols that are US EPA approved or recommended. However, most papers typically only report average/stdev for each measurement per time point/sampling location. It would be extremely helpful if this data is made available as supplementary material with the publication in tab or comma delimited format, so that the data can be easily integrated into comparative studies. On the biological side, it would be nice to have standardized sample collection, extraction and sequencing protocols across studies. It won’t take away all variabilities, but it could be a big step forward. At the same time, we felt that it is probably not the right time to do this. For some reason (…funding), drinking water studies tend to lag a little behind other fields in terms of adoption of new methods. For example, the first paper applying high-throughput amplicon sequencing to study drinking water microbial communities was more than four years after the first paper describing this method. Also, long-read amplicon sequencing is here and here, and standardizing primer choice for short-read sequencing platforms may be a moot point at this juncture. So, maybe we could standardize sample collection, DNA extraction, etc.? This is possible, but would need a critical mass of researchers in the field to come to reasonable agreement.
What seems probably easier to accomplish immediately is sharing DNA extracts between groups. It is a little challenging because of low concentrations in drinking water DNA extractions (every microliter is precious), but could be possible without making things too onerous. Probably an even easier place to start would be data sharing. We had some challenges getting access to data (and eventually only managed to get access to 66% of datasets that have been used in publications). Not all publishers make data release mandatory and in some cases the data is owned by private water utilities who are not obliged to release it. So, given that we have some way to go in terms of data openness in drinking water research, it is probably much more beneficial to focus on this in the first instance.

What I think is essential in water biodiversity studies is to know who is alive and who is dead. Based on traditional knowledge on microbial ecology, many of the detected DNA sequences in soil, dry surfaces and water belong to dead organisms, what is now called relict DNA.
Hi Maria – Thanks for reading the blog! I very much agree with you about the need to discriminate between cells that are dead and alive and that this is especially critical for drinking water systems.
As far as I can tell there are three methods currently available for discriminating between live and dead cells that also works with sequencing based approaches:
1. Cell membrane integrity: Some of the early papers on this were by Andreas Nocker and Anne Camper using Ethidium Monoazide (http://aem.asm.org/content/72/3/1997.abstract) and then propidium monoazide (http://aem.asm.org/content/73/16/5111.abstract) to selectively eliminate the signal from membrane compromised cells from mixed microbial communities. We have used propidium monoazide earlier in a drinking water setting (http://pubs.acs.org/doi/abs/10.1021/es4055725). This was the same vital stain used by the Relic DNA study by Paul Carini, Noah Fierer and co-authors (http://biorxiv.org/content/early/2016/03/17/043372.1), where they termed extracellular DNA + DNA from dead cells as relic DNA (http://biorxiv.org/content/early/2016/03/17/043372.1). Relic DNA as a term may make sense for the Carini et al study due to their sample matrix, but is likely not the accurate term in drinking water systems (or other aquatic systems) where microbial communities are not spatially static and there is a high turnover rate. Nonetheless, I am not a big fan of the term relic DNA to describe extracellular DNA + dead cell DNA because it has been used previouly in a different context (http://link.springer.com/article/10.1007%2FBF00210075). One of the key challenges while using PMA or other vital DNA stains in combination with mixed microbial communities is the protocol itself. Based on my experience in the lab with this, it takes a lot of effort/care to optimize PMA concentration and light exposure to get things right. This is especially true when sample matrices vary significantly within a study – which means that the different PMA concentrations/incubation & light exposure times may need to be used and hence, additonal controls need to be included for each methodological variation to be sure nothing goes wrong. It is a useful approach, but not always feasible in some situations (e.g. low biomass samples) or when the survey type studies where samples sizes are large. The limitation to this membrane integrity approach is that there are also cells that are dead, but still have an intact cell membrane (i.e. ghost cells). For example, in systems where UV inactivation is used – the mode of cell inactivation is DNA damage and not cell membrane damage.
2. RNA: The other approach is to extract RNA, reverse transcribe to cDNA, and then sequence cDNA. This will give information on who is active within a particular sample. This is a nice approach, but not robust way of discriminating between live and dead. Non-active does not mean dead and sometimes active (based on RNA measurements) may not necessarily mean alive (RNA, especially rRNA has a way of hanging around in inactive or even dying cells). Further, just because a cell is inactive (and thus maybe not detected using RNA approach) in a drinking water distribution system – it does not mean that the cell may not become active under different environmental conditions (e.g. the lungs or gut of a consumer).
3. Stable isotope labelling: This is one way to label DNA of actively growing cells and then separate it using neat density gradient centrifugation, followed by sequencing. This requires assumptions about what a carbon or nitrogen source a cell assimilates during growth and thus to some extent introduces a “culture-biasâ€. There is some nice work recently using heavy-water labelling followed by cell sorting (http://www.ncbi.nlm.nih.gov/pubmed/25550518) – but this will need lots more work to become accessible/high-throughput and may only be feasible for targetted questions not large scale environmental surveys.
In short, there are methods available to discriminate between live and dead cells (using a well defined metric for what constitutes a live/dead cell) – each have their advantages and shortcomings. But, none of these approaches in and of themselves will be definitive.
Which brings me to another point I have been thinking about for a while, which would be good to discuss on this forum. Is extracellular or DNA from dead cells sufficiently functionally irrelevant that we should wholesale remove it from sequencing based approaches? DNA is a biologically active molecule and can have functional implications (http://www.nature.com/nature/journal/v408/n6813/full/408740a0.html). I am not extremely knowledgeable about this side of things, but think this might be an interesting question for the microbe.net audience to engage with.
Cheers,
Ameet
Ameet and Maria,
A large group of us has been working on a review paper on this very topic that covers much of what you’re discussing and more. Hopefully to be submitted this month!
I think the question about the biological relevance of extracellular DNA is a really interesting one but I also don’t have any sense of how important it might be to the system.
Fantastic David! Look forward to reading it.