Amidst the November/December holiday chaos, myself and co-authors were proud to witness the publication of a neat new paper focused on ATM keypads in New York City. Yes, just like all other surfaces in the Built Environment, those ATM keypads are harboring lots of microbes and bits of orphaned DNA!
This ATM keypad study was work that I carried out during my previous position in Jane Carlton’s lab in the Center for Genomics & Systems Biology at New York University. The project was a collaboration between the Carlton lab and the lab of Maria Gloria Dominguez-Bello (an associate professor in New York University School of Medicine’s Human Microbiome Program). The research was partially funded by a New York University Grand Challenge project called “Microbes, Sewage, Health and Disease: Mapping the New York City Metagenome” and a grant from the Alfred P. Sloan Foundation.
This study aimed to carry out a baseline assessment of the microbes found on ATM keypads, looking at both prokaryotic (bacteria/archaea) and eukaryotic microbes using 16S rRNA and 18S rRNA amplicon sequencing, respectively. Call it “exploratory research” – we weren’t looking at any patterns in particular, because we weren’t sure what we were going to find. However, ATM keypads are an interesting micro-habitat, since they can be considered a highly trafficked surface in the Built Environment (think of how many thousands of people are probably using an average bank ATM every day in Manhattan – lots of fingers pressing those keys at all hours of the day!).
With that goal in mind, during June and July 2014 we used cotton swabs to sample microbes from 66 ATM keypads in eight neighborhoods across three New York City boroughs (Manhattan, Queens, and Brooklyn).
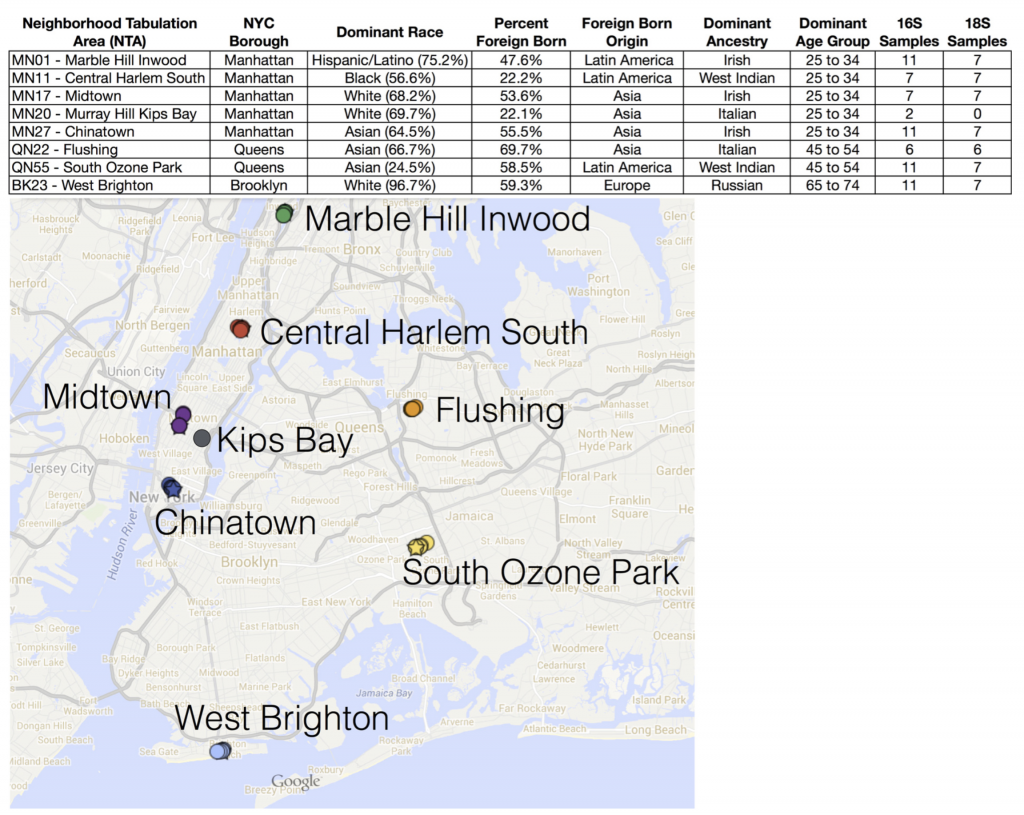
What did we find? NOTHING. That is to say – we didn’t find that microbial communities lumped together in any obvious way according to NYC geography (neighborhood or borough). We also tried to correlate microbial community patterns with population demographics obtained from the NYC Open data portal – things like age group, predominant ethnicity, etc. in each neighborhood – but again, no strong patterns there.
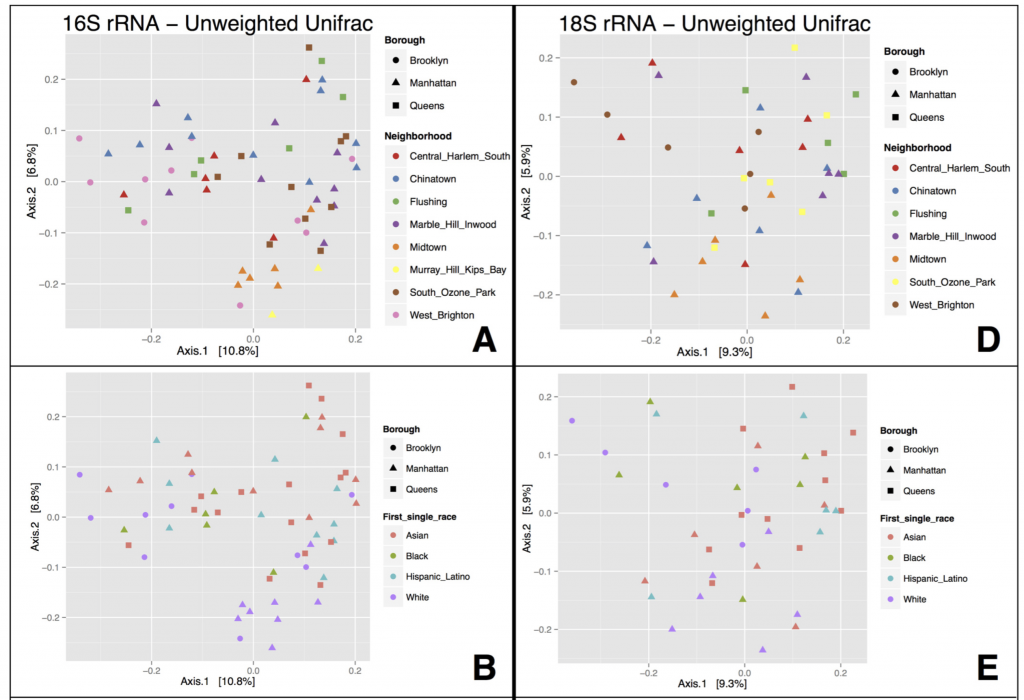
The most interesting things were the expected patterns and associations that were neat to actually see in our dataset. First, many microbes on ATM keypads seemed to be derived from the human skin microbiome – ATM microbes were similar to those found on household surfaces such as pillowcases, and TVs (surfaces which probably amalgamate an “average” of skin microbes from everyone living in the house). Other microbes on ATM keypads were similar to outdoor air (perhaps representing dust/pollen/airborne microbes settling on ATM keypads), and restroom surfaces (don’t think about that one too much…)
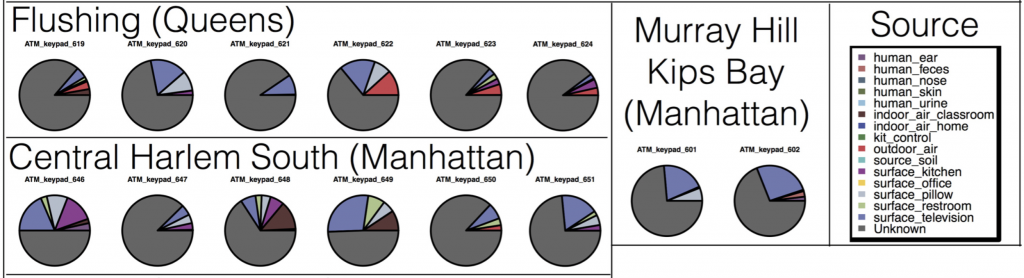
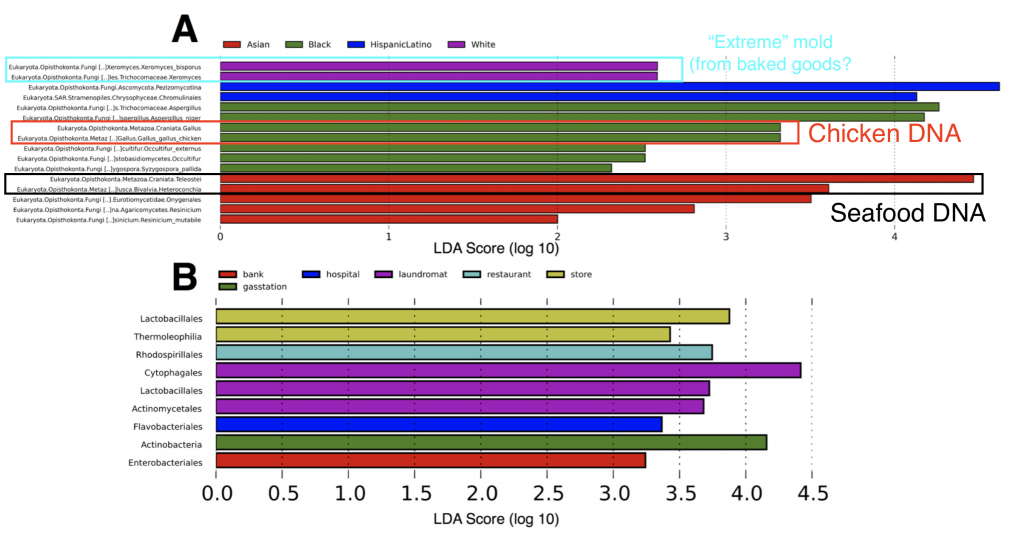
Second, in some neighborhoods we found traces of food species (chicken and seafood DNA), indicating a “microbial echo” of a person’s last meal. You might not think about it much, but when you handle food bits of DNA and cells are most likely sloughing off and sticking to your hands – so people in busy NYC may eat their meals on the go (perhaps without washing their hands) and then use an ATM keypad and transfer the food DNA onto the buttons.
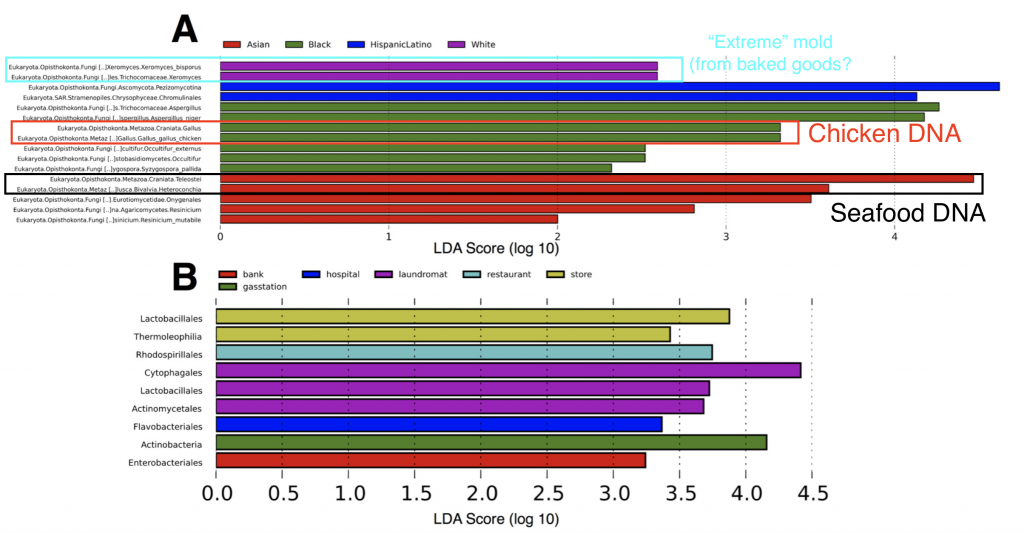
Finally we also found subtle patterns and exciting taxa in our ATM dataset, all of which are interesting topics for further research. Some ATMs showed higher abundances of specific microbial species, such as the “extreme” mold Xeromyces bisporus (the “enrichment” of this microbe was statistically significant in certain neighborhhoods – see above figure), which could potentially be used as a biomarker for fungal species associated with baked goods. X. bisporus seems to be able to tolerate low water availability and live in comparatively harsh habitats such as sugary/processed foods, hence why we can consider it an extremophile. On other ATMs we also found microbial species representing putatively parasitic taxa – Trichomonas, a sexually transmitted disease in humans, and Toxoplasmosa, the infamous parasite found in cats. While we can’t 100% confirm that these parasites linger on ATM keypads (the rRNA gene region we used does not allow us to definitely separate these parasites from other closely related protist species), these tantalizing data set the stage for future (more focused) studies of the “urban microbiome”.
If you want to hear more about this study (and more about urban microbes in general), have a listen to this radio interview I did for The Innovation Hub (a WGBH/PRI show), as part of an episode focused on “The Future of Cities”.
Reference:
Bik HM, Maritz JM, Luong A, Shin H, Dominguez-Bello MG, Carlton JM (2016)
Microbial Community Patterns Associated with Automated Teller Machine Keypads in New York City mSphere, 1(6) e00226-16; DOI: 10.1128/mSphere.00226-16
Thanks for posting!