
Back in October 2013 I wrote a blog post here called “Building science measurements in the Hospital Microbiome Project: Part 1” where I described the types of building environmental and operational measurements we were making at the time as part of Jack Gilbert’s Sloan-funded Hospital Microbiome Project (Jeff Siegel at the University of Toronto also played a major role in these measurements and in bringing our team on to the project). Fast forward to present day and we’ve since collected all of our data — we actually finished data collection about a year ago — and published a paper in PLoS ONE, “Spatial and temporal variations in indoor environmental conditions, human occupancy, and operational characteristics in a new hospital building,” which is available online now. Here I will briefly summarize our key findings.
As a quick reminder, our team worked to measure a variety of indoor environmental conditions, human occupancy, and operational characteristics in 10 patient rooms and 2 nurse stations across 2 floors of the hospital, which we hypothesize may help explain some of the differences in microbial communities that are observed by Jack’s team over the course of the sample year. Building science measurements included environmental conditions (indoor dry-bulb temperature, relative humidity, humidity ratio, and illuminance) in the patient rooms and nurse stations; differential pressure between the patient rooms and hallways; surrogate measures for human occupancy and activity in the patient rooms using both indoor air CO2 concentrations and infrared doorway beam-break counters; and outdoor air fractions in the heating, ventilating, and air-conditioning systems serving the sampled spaces. We also utilized passive sampling of airborne microbes using a thin sheet of HVAC filter media installed over the return grilles in the rooms.
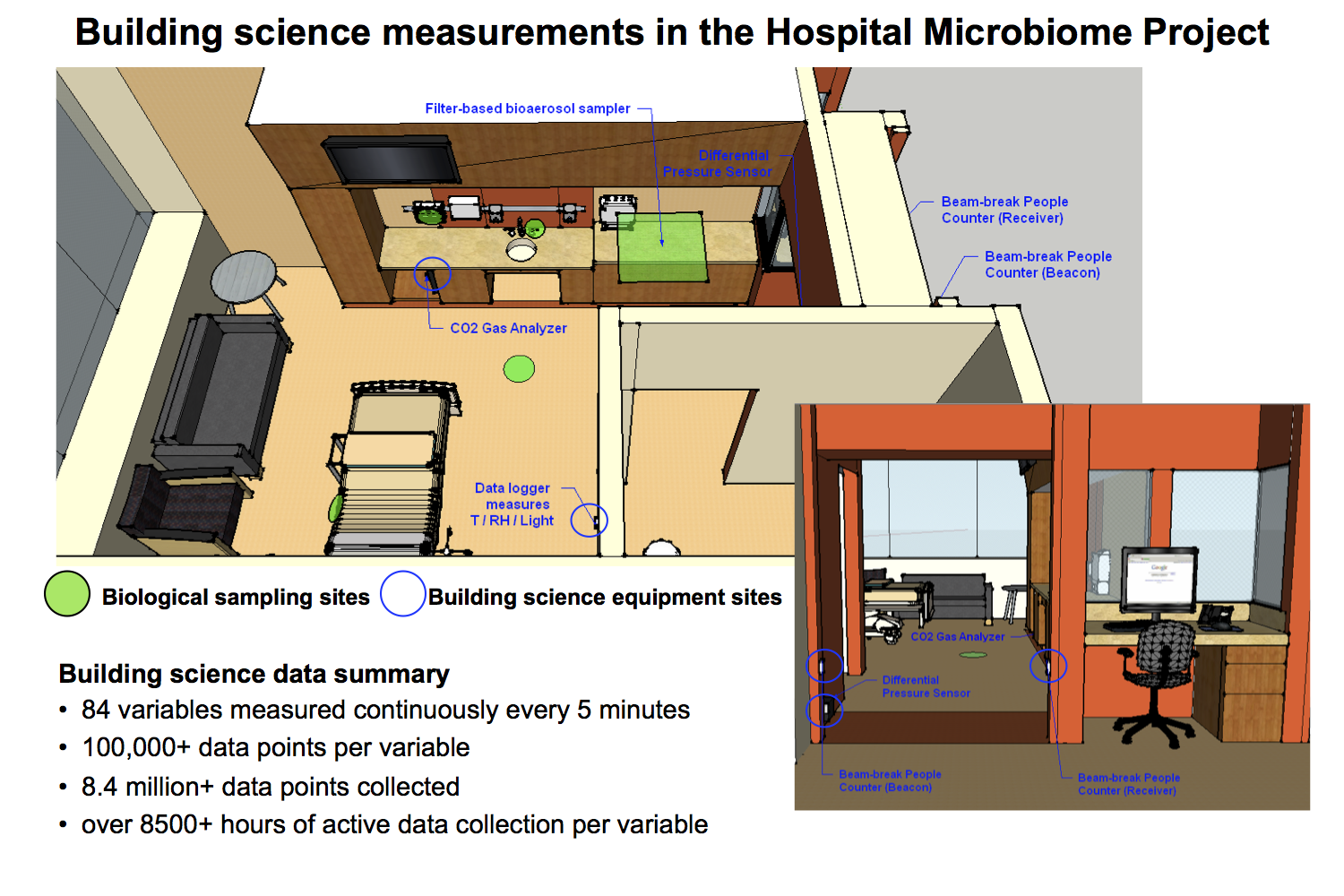
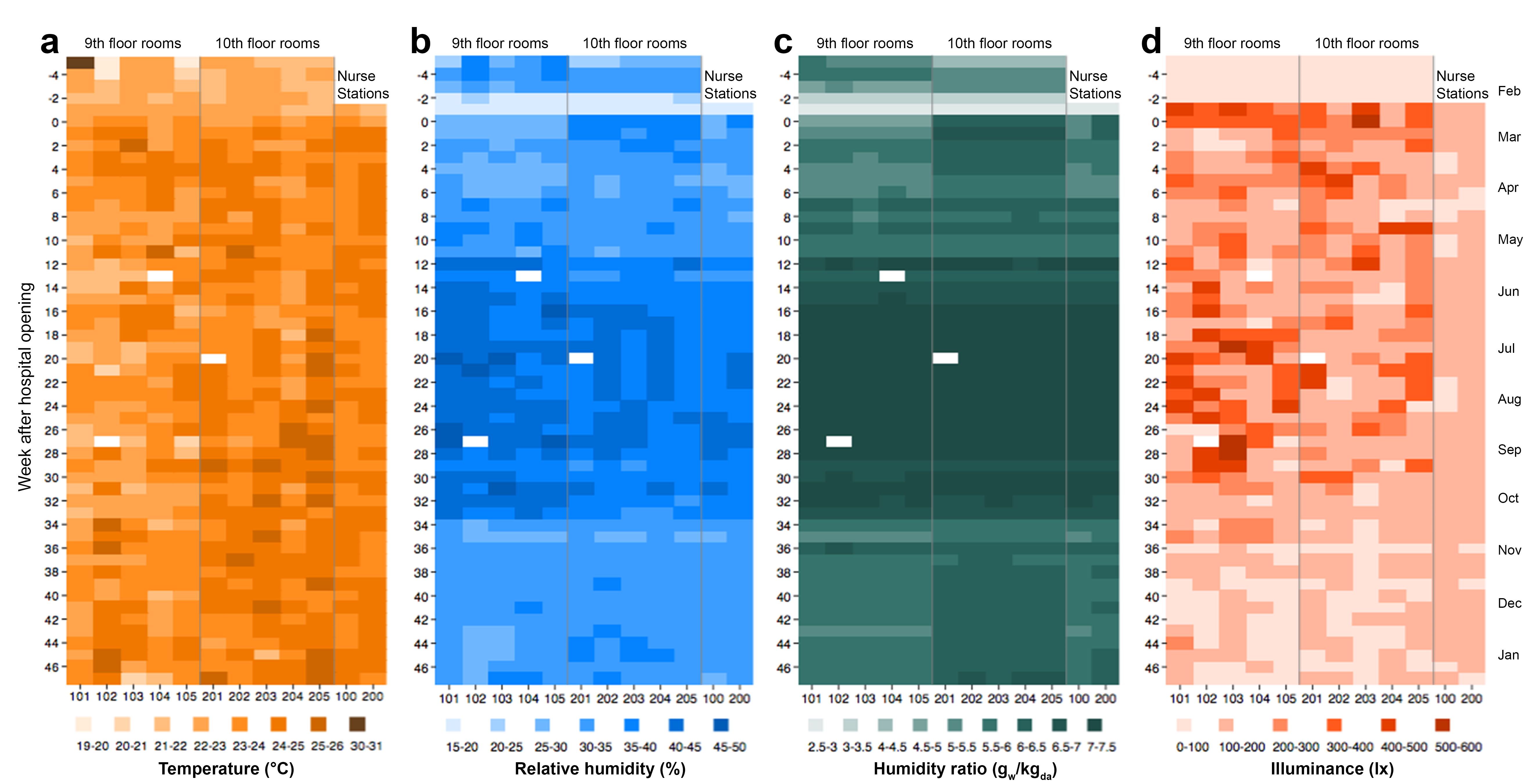
The data began pouring in February of 2013 and we finished collection in mid-January of 2014. At the end of the nearly yearlong monitoring period we ended up with approximately 100,000 time-series building science data points associated with about 80 different variables throughout the hospital and patient rooms for a total of over 8 million data points! This was a HUGE built environment data collection effort — one of the largest long-term building science data collection campaigns that I’ve seen. To give you an idea of how much and what kinds of data were collected, I’ve provided a figure below showing weekly averages of environmental conditions in the patient rooms and nurse stations measured over the duration of the project.
Briefly, the main findings from this work include:
- Indoor temperature, illuminance, and human occupancy/activity were all weakly correlated between rooms, suggesting that occupants of otherwise very similar rooms maintained control of their indoor temperatures and lighting levels, and people entered and exited their rooms in very different patterns of intensity
- Relative humidity, humidity ratio, and outdoor air fractions showed strong temporal (seasonal) patterns and strong spatial correlations between rooms, suggesting that humidity and ventilation rates were largely governed by the mechanical systems serving each floor and did not vary from room to room
- Differential pressure measurements confirmed that all patient rooms were operated at neutral pressure, per the hospital’s design specifications
- The patient rooms averaged about 100 combined entrances and exits per day, which suggests they were relatively lightly occupied compared to higher traffic environments (e.g., retail buildings) and more similar to lower traffic office environments; however, there was a broad distribution in these data
- There were also clear differences in several parameters before and after the hospital was occupied with patients and staff: air temperatures, relative humidity, and occupant activity were all higher after the hospital opened
You can also find a blog post about this project on my Built Environment Research Group website and on the Hospital Microbiome Project website. It’s been a pleasure working on this project — stay tuned for future publications coming from this project, including results from the DNA sequencing efforts.