Last week, I came across a paper in PLOS ONE that looked interesting, especially in the light of the recent mBio paper that looked at sewage as a reflection of a city’s human-associated microbiome (also see this recent post on MicroBEnet).
In the PLOS ONE paper “The Source of the River as a Nursery for Microbial Diversity“, de Oliveira and Margis investigated the microbial composition of the Sinos River in Brazil. Rio Dos Sinos is roughly 200 km long and consists of three parts: a rapidly flowing upper course with a low population density, a middle course running through rice crop lands with increasing amounts of urbanization, and a slow flowing, more polluted, lower course that runs through Sao Leopoldo City.
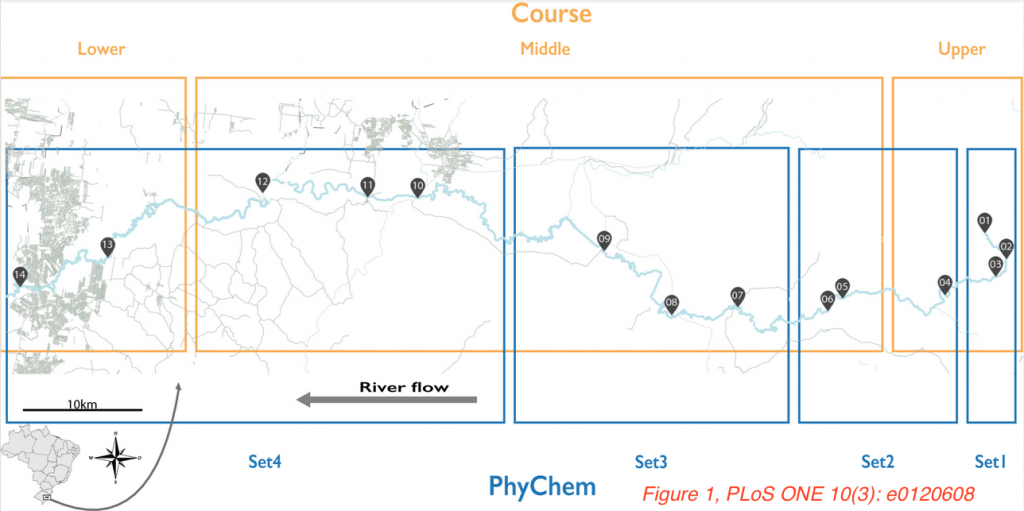
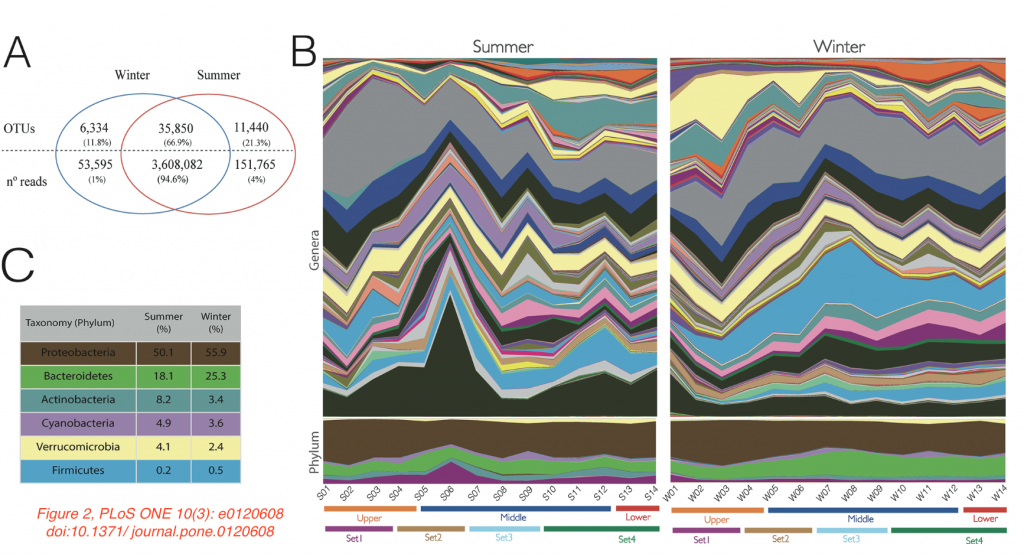
The authors wanted to investigate the influence of urbanization and pollution on the microbiome composition of this river. They sampled 14 different sites along the three parts of the river, both in the summer as well in the winter, extracted nucleic acids, amplified and sequenced the V4 region of the 16S rRNA gene. Among 5.7 million reads, over 50,000 bacterial and archaeal OTUs were found. Proteobacteria, Bacteroidetes, Actinobacteria, Cyanobacteria, Verrucomicrobia and Firmicutes were the most abundant bacterial phyla.
Interestingly, despite the different flow and pollution rates, the river’s microbiome was relatively stable over the different sampling sites and in different seasons. All water samples contained a core group of OTUs, with the OTUs present in the sample taken closest to the river’s origin representing 94% of the reads in all samples. However, statistical analyses (mantel test, Adonis, ANOSIM), showed significant differences between samples taken from different parts of the river or different seasons, but there was no clear relationship between the level of urbanization at different sites of the river and microbiome composition. Of note, the different sites were more similar to each other in the winter, which according to the authors could be explained by increased rainfall diluting and mixing the water.
Although this is maybe a small paper without any big conclusions, I liked the concept of following a stream of water over time and location. Or maybe it is just my love for maps?
The paper has some flaws too. The 16S rRNA gene is consistently called “16S” (booh!), and there is an error in the amount of bacterial phyla found (40 instead of “4” mentioned on page 5). There also appears to be an error with the colors in Figure 2B and C (see image above; the yellow Verruco’s in C might correspond to the pink areas in the lower part of B). I would also like to have seen more information about each location’s treatment of sewage before its into the river. And finally, a statement about the deposition of the sequences in a public database appears to be missing.
Despite these errors (which should have been caught during peer review), I thought this was a nice small study that deserved to be highlighted in the ever increasing flow of microbiome papers.
The Source of the River as a Nursery for Microbial Diversity – Luiz Felipe Valter de Oliveira, RogeÌrio Margis
PLoS ONE 10(3): e0120608. doi:10.1371/ journal.pone.0120608
We’ve tried this for the River Thames, back in 2008 and in view of the London Olympics. Never funded. For the record.
@Christos, That sounds like a great project, esp. with the Olympics in mind. Too bad you did not get the funding. That unfortunately affects many of us and many of our great ideas.
I keep hearing this objection, and I have to admit I don’t understand – what is the objection to using “16S” as shorthand for the 16S rRNA gene?
@Rachel: I will immediately agree with you that “16S” is so much shorter and easier to use than the full name, and many scientists will call it like than when they talk to each other. But in scientific publications, we try to be more precise in naming things, so there we use “16S rRNA gene”, which is indeed quite a mouthful. If you would only say “16S” you would technically refer to a Svedberg unit, which is a sedimentation rate during ultracentrifugation. Instead, we want to refer to the gene encoding the ribosomal subunit that has that particular sedimentation rate. So it is just a matter of carefully wording what we refer to in a scientific paper.