 Gearing up the UNITE database for the built mycobiome
Gearing up the UNITE database for the built mycobiome
The team behind the UNITE database for molecular identification of fungi has been granted support from the Sloan Foundation to strengthen the support for fungi from the built environment. Launched in 2001 as an ITS database for identification of ectomycorrhizal fungi in the Nordic countries, UNITE has since grown into a full-fledged sequence management environment covering all fungi at a global level. It mirrors GenBank and the INDSC and features all public fungal ITS sequences. UNITE supports and encourages third-party sequence annotation, and several annotation efforts have brought the data up to standards for, e.g., plant pathogenic fungi, mycorrhizal fungi, and various parts of the Dikarya. No annotation effort has however covered the mycobiome of the built environment in an explicit way.
The funding granted will be used for three overarching tasks:
- Organize two sequence annotation efforts. One of these will bring together taxonomic experts on building-related fungi to go through the taxonomic annotations and circumscriptions of public sequences and species from the fungal lineages of their expertise. The second workshop will restore missing metadata for all building-related fungal sequences following the MIxS-Built Environment standard.
- Sequence type material from building-related fungi and make the sequence data (ITS and nLSU) available as formal reference sequences through UNITE and the INSDC (and for the downstream initiatives that rely on UNITE for reference sequences). The focus will be on hitherto unsequenced type material.
- Develop UNITE to better support the built mycobiome. We will take various measures to improve the support for building-related fungi in UNITE, such as implementing support for the MIxS-Built Environment standard, establishing a taxonomic feedback loop whereby potentially new species of building-related fungi are brought to the attention of relevant taxonomists, and adding support for non-fungal organisms often found in the built environment.
Various other tasks, such as examining how commonly used pipelines and software environments can be improved to better account for the built mycobiome, will also be performed. To increase the interaction with researchers and research professionals in fields relating to the built environment, the UNITE team will attend and present our initiative at relevant conferences and meetings in these fields.
We will do our best to use the funding in a way conducive to molecular identification of building-related fungi. All our results will be open, free, and available to the scientific community at no delay. We similarly hope that the increased visibility of UNITE in building-related contexts will generate a momentum for community participation in the annotation and data sharing efforts. All contributions are most welcome.
Andy Taylor, University of Aberdeen (formal PI, Andy.Taylor@hutton.ac.uk)
Urmas Kõljalg, University of Tartu (co-PI, urmas.koljalg@ut.ee)
Henrik Nilsson, University of Gothenburg (contact person, Henrik.nilsson@bioenv.gu.se)
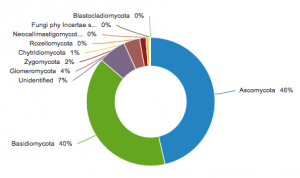
Figure 1. Distribution of UNITE species hypotheses across fungal phyla.
Congratulations to the UNITE people. You are doing a very valuable work in combining several methods in order to clarify fungal taxonomy