(This post was written by group#1 as a writing assignment)
A new class yet to be named begins giving students hands on experience with genomic sequencing.
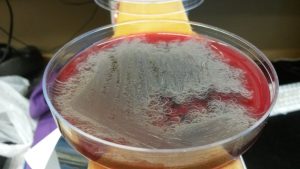
We are attempting to sequence bacteria from diseased abalone. We had fecal samples from both healthy and diseased red abalone and white abalone, 4 in total. In the first day of our class, we separated bacteria to sample. Each group has 4-5 people who each grabbed 2 bacteria from colonies that had grown on different types of agar plates, hopefully resulting in 8-10 different bacteria from each abalone. We dilution streak plated the bacteria in an attempt to isolate colonies. In the following weeks, we will perform PCR to get a broad idea of which bacteria we chose. Hopefully, we will find a bacteria worth sequencing. The importance of sequencing the genome of bacteria growing in Abalone feces is relevant because of the increasing impact of Abalone withering syndrome, which is mainly transmitted through fecal matter. Withering syndrome has had growing impact on Abalone population since 1985 and their endangerment not only affects the food industry, but also their delicate ecological habitat in which they are fundamental.