“Don’t tell me what you found.”
“Oh my gosh it must be disgusting!”
“Can people transfer vaginal microbes to each other?”
These are some of the questions we’ve gotten from subway riders and press. Despite these reactions, our study on the Boston subway system is now out on MSystems! Recently, there has been a rise in culture-independent subway studies. Subway aerosols were profiled by Robertson et al in New York, Leung et al in Hong Kong, and Dybwad et al in Oslo. Just last year, Afshinnekoo et al characterized subway surfaces. The initiative he is leading with Chris Mason and multiple colleagues, MetaSUB, aims to sample subways around the world.
The Boston MBTA project originally began in 2013 with Kasia Baranowski (a rotation student) and Xochitl Morgan (a research associate) in Curtis Huttenhower’s lab. Our goals were to figure out 1) what microbial communities were on subway surfaces, 2) how material, location, and traffic affected them, and 3) whether these communities served as reservoirs for pathogenic potential. Kasia and Xochi started out testing whether Copan floqswabs or cotton swabs gave higher yields, as well as whether and which swabbing solutions should be used. They then passed the project on to me (an incoming rotation student) and Regina Joice (a new postdoctoral fellow). In the Huttenhower Lab (hutlab), we have a buddy system. New students are paired with postdocs, and new postdocs are paired with senior postdocs or research associates. Even though we were both new members, when I first got to the hutlab, Regina was my buddy, and Xochi was Regina’s buddy.
I am thankful that we had this system. To me, this project was extremely exciting but completely un-”controlled”. As an undergraduate and technician, I had only worked with model organisms and their select genes. How were we going to sample the “T” (the local name for MBTA) with so many uncontrolled variables? Get enough DNA? Deal with variation in weather and different train cars? If someone just got off a seat – should we sample it or only go for empty seats? Together with Regina, Kasia, and Xochi, we tried to create some structure out of the chaos. Eventually, Regina implemented a sampling strategy to collect samples from 6 surface types on trains, including seats, seat backs, grips, horizontal poles, vertical poles, and the walls, which would be a type of control since it should be infrequently touched. We also sampled 2 surface types inside stations, including touchscreens and the sides of ticketing machines. Regina mapped out which stations and trains we would ride beforehand, and we went on multiple test runs to make sure we could carry it out.
On sampling trips, we always went with Jose Vallarino from Jack (John D.) Spengler’s lab as well as an MBTA police escort. Jose was a key part: for train sampling, he would park his car at a central location (Park Street or South Station), with the windows vented and a box of dry ice. We dropped off samples between transfers to different lines, so no sample had to sit on “plain ice” for too long. For touchscreen sampling, he drove us to the outermost stations. To keep things consistent, Regina always swabbed (only 1 or 2 of my swabs made it in), while Jose assisted by handling the swabbing solution, tubes, and any trash generated, and I took pictures and notes.
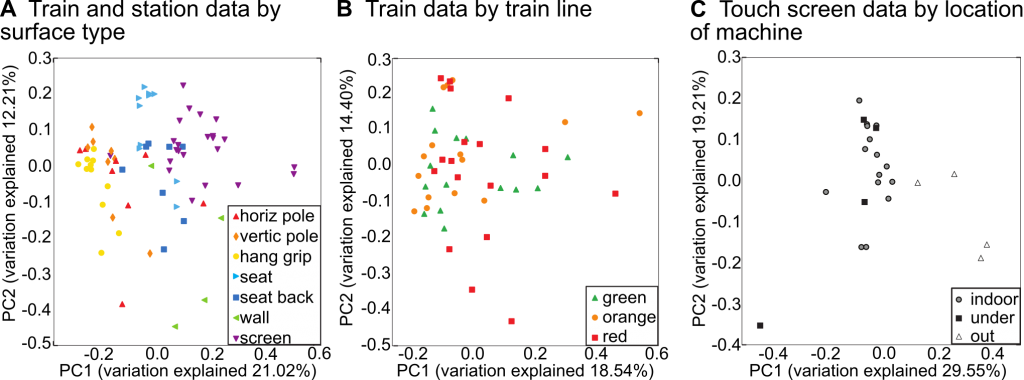
In the end, it was this design that helped make the Boston subway study unique. We were able to look at how microbial communities changed between surface type, surface material, and geography using both 16S sequencing (of the V4 region) and shotgun sequencing. Through ordinations of the 16S data, we saw that surface types mattered greatly, along with indoor/outdoor location for ticketing machines. Surprisingly, geographic variation did not matter much, but this could be due to Boston’s small size and the T’s dense and diverse ridership.
The 16S and shotgun results were mostly complementary. Both showed high abundances of Staphylococcus, Corynebacterium, and Streptococcus. The main difference was that the V4 primers could not detect Propionibacterium acnes: these primers are not ideal for skin microbial communities – which is what the subway most resembled (based on Microbial SourceTracker and correlations in abundance with skin taxa). This provided an interesting contrast of the strengths and weaknesses of amplicon versus shotgun alignment-based approaches. Metagenomes showed that P. acnes had a mean abundance of 47% in the population. In the same data, the most abundant viruses present were phage that corresponded with some of the most abundant genera.
The use of shotgun sequencing also allowed us to quantify antibiotic resistance genes from the Comprehensive Antibiotic Resistance Database (CARD), as well as virulence genes from the Virulence Factor Database (VFDB). Overall, we found very low numbers of these genes: at least for antibiotic resistance genes, there were less than you’d find in the human gut. We’re now moving onto PMA-based assays to see what is alive and dead on the subways. These are first steps towards assessing how microbial profiling might be useful for biosurveillance, which may come in the form of assessing specific taxa, monitoring indicator taxa with co-occur with pathogens, or quantification of pathogenicity genes.
Even with this publication, I guess thoughts of the train will never leave us. In my June trip to the Twin Cities, all I could think about was how clean and quick their Green Line was as I went from Minneapolis to St. Paul. And Regina? She’s getting married at a train station come August!
One thought on “Microbes on the Boston “T””