In addition to thinking about appropriate statistical analyses given the compositional nature of microbiome data sets (see the previous post here), there is a related but different issue that’s back on my mind – the benefit that quantitative information would bring to research questions. My engineering colleagues in the built environment world have consistently reminded me (read: nagged) that interpretation of microbiome data sets will be limited for certain questions until quantity is part of the data.
On this topic, there’s a new paper out describing quantitative microbiome profile in the context of the human gut. The authors use flow cytometry to determine cell counts per gram of substrate, and use that information, along with an operon copy number correction, to generate a Quantitative Microbiome Profiling (QMP) matrix.
While the approach is different, the goal is the same one that motivated Karen Dannemiller et al. to combine qPCR with high throughput sequencing to inform estimates of human fungal exposure in aerosols.
The particular substrate of interest (poop versus aerosols – may the twain never meet) will likely determine the best method for determining biomass, but it’s exciting to see that there is growing interest in the need for quantity-informed microbiome data sets – and maybe it will spur innovation in this area.
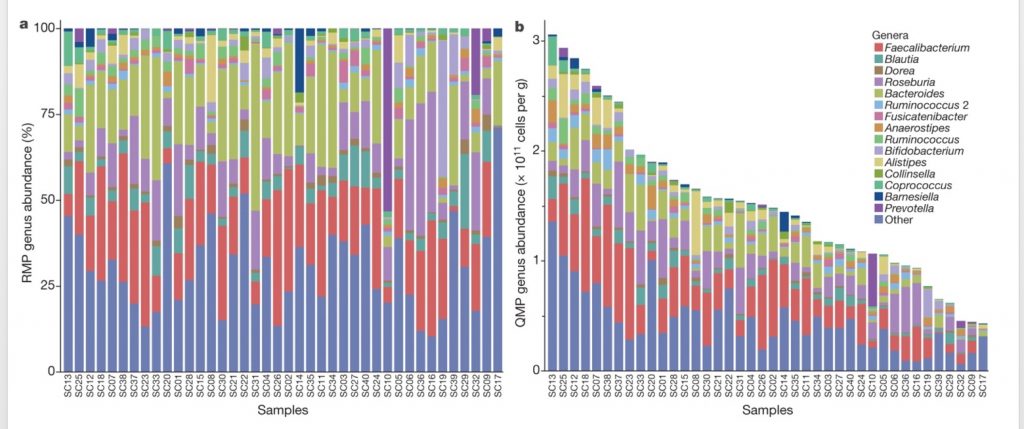
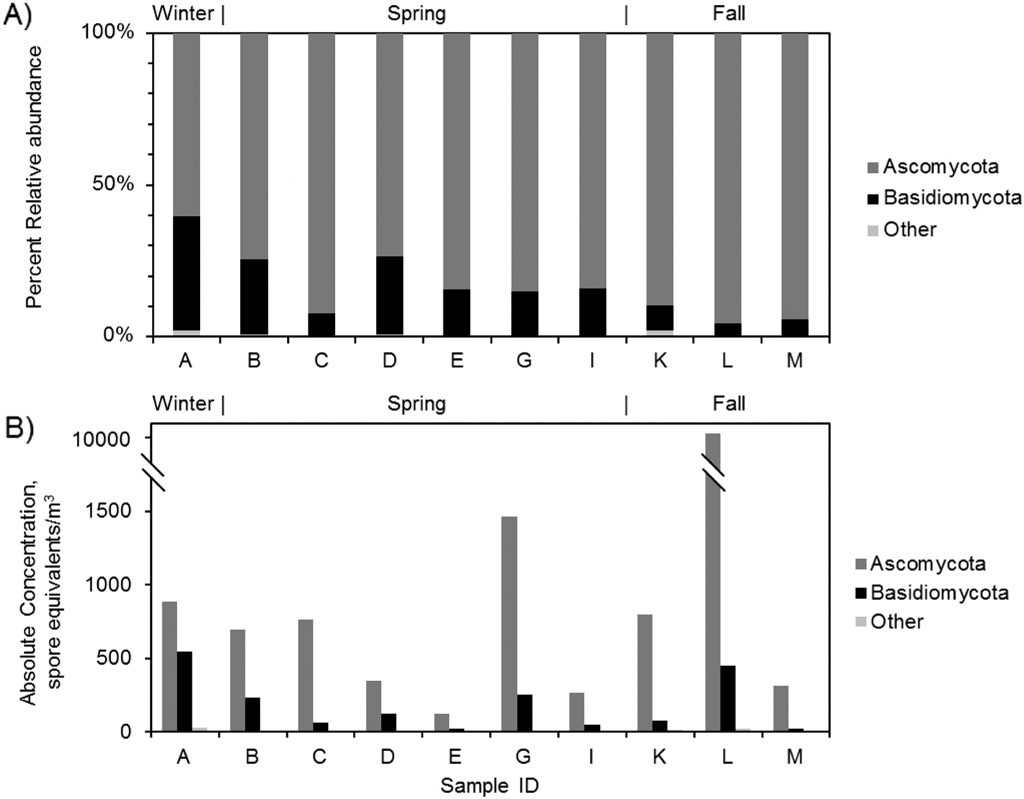
Yes! We want to be more quantitative! Regarding “poop versus aerosols,” there are unfortunately many places where the twain meet: toilet flushing, aeration basins at wastewater treatment plants, land application of sludge, and more. We considered this possibility at length in our work on the potential for aerosolization of Ebola virus.
Great post, thanks Rachel. There are still many other systems that could benefit from quantitative measurement. One challenge might be determining the best denominator in a field like dentistry. Bacterial genomes per gram/mL of saliva? Or per mouth? Curious to hear what others think.