New paper of interest:: Genomic Analysis of Hospital Plumbing Reveals Diverse Reservoir of Bacterial Plasmids Conferring Carbapenem Resistance
Weingarten RA, Johnson RC, Conlan S, Ramsburg AM, Dekker JP, Lau AF, Khil P, Odom RT, Deming C, Park M, Thomas PJ, NISC Comparative Sequencing Program, Henderson DK, Palmore TN, Segre JA, Frank KM. 2018. Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio 9:e02011-17. https://doi.org/10.1128/mBio.02011-17.
I found out about it on Twitter and am just going to embed the Tweets I read which led me here since they are useful:
Next time you pass a hospital, consider the “vast, resilient reservoir” of antibiotic resistant bacteria trickling through pipes beneath your feet. https://t.co/alFWfhMBmt
— Jamie Hall (@jpjhall) March 22, 2018
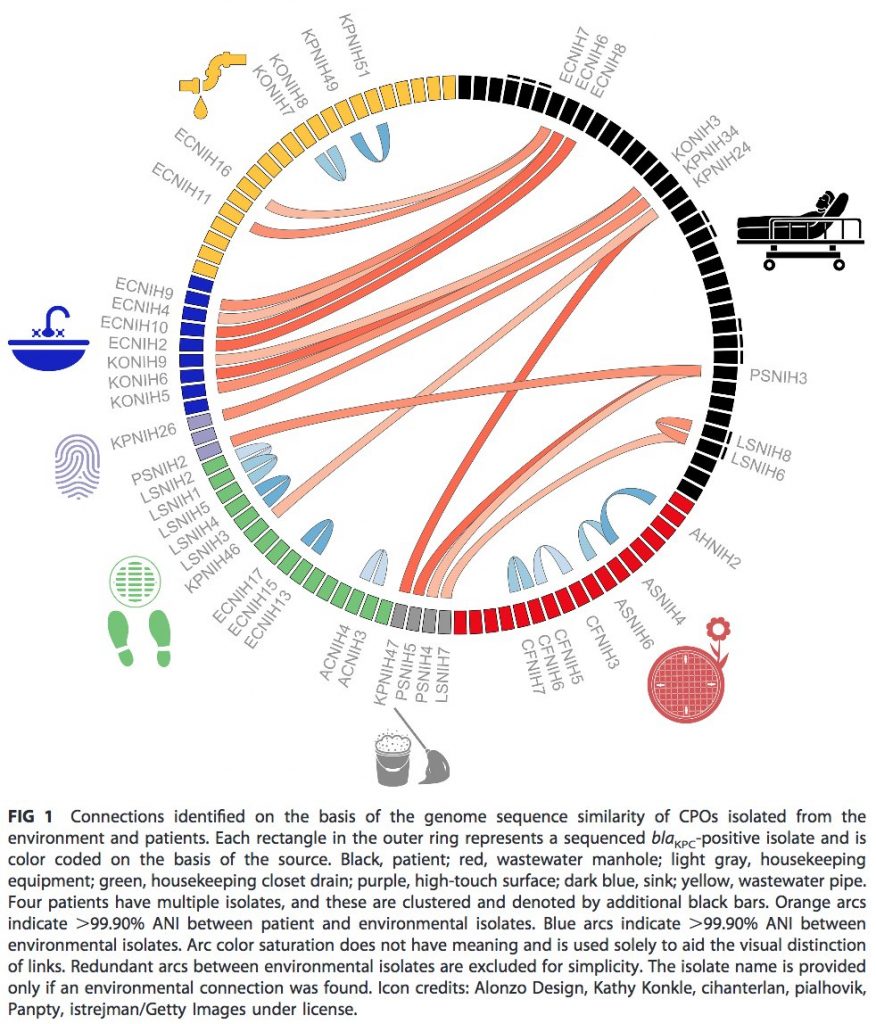
In this @mBiojournal paper, genome surveillance of patients, drains, mops, and manholes showed hospital wastewater is full of antibiotic resistance plasmids. pic.twitter.com/ODMhONevVy
— Jamie Hall (@jpjhall) March 22, 2018
This doesn’t just reflect the bacteria that patients are carrying. Wastewater often carried resistance elements that were rare in patients. pic.twitter.com/p3IdcqByFL
— Jamie Hall (@jpjhall) March 22, 2018
The high resolution sequence data combined with patient records even allowed inference of resistance elements recombining between plasmids in a hospital sink drain. pic.twitter.com/H5gY0M5bEp
— Jamie Hall (@jpjhall) March 22, 2018
Hospital sinks are interesting places for a microbiologist — potentially the main meeting place of clinical and environmental bacteria. https://t.co/MWhhZZjPA8
— Jamie Hall (@jpjhall) March 22, 2018