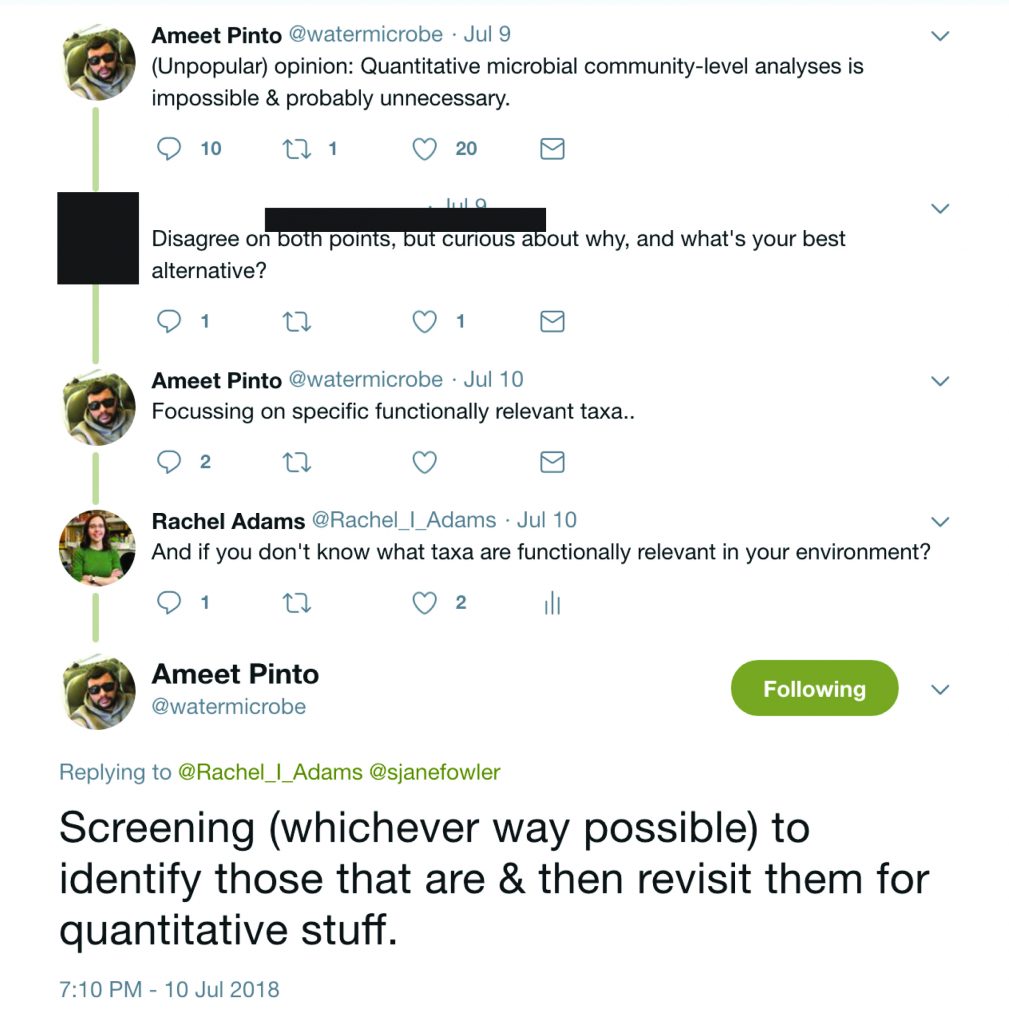
One of the hurdles in linking microbial ecology with building science has been incorporating quantitative information about the microorganisms encountered in indoor environments, mainly because the standard high-throughput amplicon approach for community analysis is semi-quantitative, at best. Over the summer, there was a Twitter conversation related to this topic.
My take-away from this (what I view as a productive) Twitter exchange is the idea that community analysis is one way to identify the important taxa and generate hypotheses. Those relevant taxa would then be further explored with, for example, qPCR. Seems like a solid approach to me. The hard part, of course, is in identifying the functionally relevant taxa that should be revisited with quantitative approaches. Which brings me to this year-old paper:
Rosen, Connor E., and Noah W. Palm. “Functional classification of the gut microbiota: the key to cracking the microbiota composition code: functional classifications of the gut microbiota reveal previously hidden contributions of indigenous gut bacteria to human health and disease.” BioEssays 39.12 (2017): 1700032.
The authors detail many reasons why, outside of infectious disease and satisfying Koch’s postulate, identifying the microbiota responsible for the disease state is challenging. Once a particular function or trait has been identified (they use the example of the production and secretion of immunoglobulin A), they discuss a new approach, relying on fluorescence-based cell sorting followed by 16S gene amplicon sequencing, to isolate those taxa associated with that trait.
In applying this approach to the built environment, the challenge (and opportunity!) comes from identifying specific microbial traits or functions that we’d like to either promote (e.g. those present in farming environments which protect respiratory health) or minimize (e.g. those present in water damaged buildings which are associated with ill respiratory health). Then quantity can also be incorporated to create a more complete picture of the roles that environmental indoor microbiota play in occupant and building health.
I suggest to use other methods than sequencing to document taxa that are functionally relevant in the building environment. Many microbes can neither grow nor reproduce indoors. They are only accidentally present as “backgroud noise” in the building environment. However, they cope many positions in the long OTUs lists obtained by sequencing. Traditional microbiologists have spotted and characterized many of them: mycorrhiza fungi, animal specialists, parasites, the majority of anaerobes, many hardwood specialists… Although they can be identified by culturing and microscopy, many of these species are not sequenced yet, are assigned to higher taxonomic levels as family or order, or worse, they are incorrectly identified by DNA sequencing.
Functional microbes indoors, aka indoor key species, are those that can grow and reproduce in buildings, and interact with each other and building substrates. A simple tape samle taken on building surfaces and observed with an optical microscope will reveal which fungi are growing and reproducing on the spot. The reproducing structures generated on building substrates allows us to identify them without culturing in the lab. If you grow these samples in the lab, you may force dormant species to artificially reproduce, and wrongly conclude that they are active on the sampling place.
So please, use us traditional taxonomists. We have knowledge – and publications – on building ecology that can be useful for those working on DNA.
Of course those microbes that can grow and reproduce in buildings are functionally relevant, but they are just one component of the functionally-relevant indoor microbiota. There are many other examples of research in which you are not looking at moldy building substrates. For example, during building investigations when the question is whether there is hidden mold, so you take air or vacuum samples. Or when comparing microbes in the house dust of urban and farm environments and linking those with respiratory health. But I agree with you that understanding which taxa can actually grow and reproduce on buildings is an important part of the picture, and knowledge from ‘traditional taxonomists’ on building ecology is invaluable.