Recently, I have been reading and thinking a lot about what one needs to develop a model system for host-microbiome studies. I am particularly interested in this because I have been working with a few colleagues on developing seagrass, and specifically, Zostera marina, into such a model system. This started with a grant from the Gordon and Betty Moore Foundation to start the “Seagrass Microbiome Project” a few years ago and has continued with various efforts since.
More about that another time. What I want to do here is just to share the some notes on articles I have been reading about this. I am trying to include open access papers here as much as possible. Note – I have put text from these papers in quotes.
I would love other examples of papers or blog posts or talks or books or such on this topic and of course comments too …
Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, et al. (2017) Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol 15(3): e2001793.
- Note – I am a coauthor.
From the abstract: “Here, we identify priorities for research in this area:
- (1) develop model host–microbiome systems for crop plants and non-crop plants with associated microbial culture collections and reference genomes,
- (2) define core microbiomes and metagenomes in these model systems,
- (3) elucidate the rules of synthetic, functionally programmable microbiome assembly,
- (4) determine functional mechanisms of plant-microbiome interactions,
- (5) characterize and refine plant genotype-by-environment-by-microbiome-by-management interactions.”
Existing and developing model systems
- “A suite of model plant species is required to understand which processes will translate to crops at varying evolutionary distances [41]. Progress has been made towards the establishment of model host–microbiome systems for the legume Medicago [42], Populus [43], rice [13,15,44–46], Sorghum [47], Miscanthus [48], maize [49,50], and tomato [51].”
- “All of these model organisms have fully sequenced genomes and growing communities of researchers to extend their utility for microbiome research.”
- “Coordinated efforts to establish public resources, such as repositories and curated databases for sequenced culture collections of associated microbiota, are necessary for full maturation of model systems.”
What is needed in ag systems
- Field microbiome studies: “In addition to model systems and culture collections, targeted studies of agricultural microbiomes in the field provide key foundational knowledge that can lead to innovation in several ways.”
- Rules of community assembly: “Beyond knowledge of the identity and functional attributes of microbes present in crop plant “core” microbiomes, determining the rules by which microbes assemble into those communities will be essential for any attempt to manipulate or manage the agricultural microbiome”
- Functional mechanisms
- Plant genotype × microbiome × environment × management interactions
Wilkins LGE, Leray M, O’Dea A, Yuen B, Peixoto RS, Pereira TJ, et al. (2019) Host-associated microbiomes drive structure and function of marine ecosystems. PLoS Biol 17(11): e3000533.
- Note – I am a coauthor.
- Just published – focuses on marine host associated microbiomes
- Argues that a model system would be a location / ecosystem and not just a specific host
Chevrette MG, Bratburd JR, Currie CR, Stubbendieck RM. 2019. Experimental microbiomes: models not to scale. mSystems 4:e00175-19. https://doi.org/10.1128/mSystems.00175-19.
- argue for the importance of reductionistic approaches “Reductionist experimental models are essential to identify the microbes involved in interactions and to characterize the molecular mechanisms that manifest as complex host and environmental phenomena.”
- focus on three models (Bacillus-Streptomyces, Aliivibrio fischeri-Hawaiian bobtail squid, and gnotobiotic mice
- Most useful in terms of host-microbiome studies regarding discussion of gnotobiotic systems
- “Overall, gnotobiotic animals provide an approach to interrogate the role of complex microbiota in emergent phenotypes of interest by reducing the complexity to controllable independent variables (e.g., a single bacterial strain or product).”
- “delineating community states that contribute to emergent properties and complex interactions will require experimental models, and the ideal balance between a model’s complexity, ease of manipulation, and overall biological relevance will depend upon the scientific questions posed“
Rodriguez PA, Rothballer M, Chowdhury SP, Nussbaumer T, Gutjahr C, Falter-Braun P. Systems Biology of Plant-Microbiome Interactions. Mol Plant. 2019 Jun 3;12(6):804-821. doi: 10.1016/j.molp.2019.05.006. Open Access.
- “To unravel the complex network of genetic, microbial, and metabolic interactions, including the signaling events mediating microbe–host interactions, comprehensive quantitative systems biology approaches will be needed“
- Synthetic communities
- “A powerful approach to study complexity in a controlled setting is the use of bacterial SynComs (Table 1)”
- “An exciting study toward understanding cross-kingdom interactions was reported by Duran et al. (2018) studying the A. thaliana root microbiome. After profiling bacteria, fungi, and oomycetes, they established microbial cultures for all three groups to investigate their interactions.”
- Metabolites
- “Among the fundamental principles of microbiome–host interactions are metabolic exchanges“
Establishing Causality: Opportunities of Synthetic Communities for Plant Microbiome Research. https://doi.org/10.1016/j.chom.2017.07.004
- From abstract:
- “Here, we discuss reductionist approaches to disentangle the inherent complexity of interactions in situ. Experimentally tractable, synthetic communities enable testing of hypotheses by targeted manipulation in gnotobioticsystems. Modifications of microbial, host, and environmental parameters allow for the quantitative assessment of host and microbe characteristics with dynamic and spatial resolution.”
- Experiments needed
- “While generating catalogs of sequencing data through various methods (e.g., 16S rRNA gene-amplicon sequencing or whole-genome shotgun metagenomics) is readily accomplished, elucidating the biological mechanisms involved in microbial community assembly, dynamics, and resilience/resistance, as well as establishing causal relationships between the microbiota and plant phenotypes, is not clear-cut.”
- Overview of what experiments could be done
- “Addressing such fundamental principles is challenging and will require experimental approaches that allow for reproducible conditions and targeted modifications of selected factors in order to identify and link changes at the genetic or molecular level to host and community phenotypes (Figures 1 and 2).“
- Microbial factors that could be tested include
- the presence/absence and abundance of particular microbes or groups
- the order in which various microbes are introduced
- genetic alterations of particular strains or the host, and growth conditions.
- Modifiable environmental factors include
- soil type
- temperature
- humidity
- intensity and quality of light,
- biotic factors such as insects.
- “By utilizing appropriate containment and targeted manipulation techniques, the inherent complexity provided by the genotype of the host, the genotype of the microbiome, and the environment (GH× GM × E) can be reduced in order to establish causality“
- Emphasizes that both top down (working with natural communities) and bottom up (synthetic communities) are beneficial.
- Synthetic communities
- “Translated to the plant microbiota, a synthetic community or constructed microbiome is a microbial community designed by mixing selected strains using bottom-up combinations and applying it to plants to study various aspects of plant-microbe interactions with the aim to uncover fundamental principles and generate knowledge that can be translated into agricultural applications (Figure 2).”
- Has detailed protocol on how to select synthetic communities “Synthetic communities are built bottom-up from individual microbial cultures and allow the dissection of the roles played by distinct strains in higher-complexity interaction networks (see above).
- Culture collections and genomes
- “Comprehensive culture collections of microorganisms are a prerequisite for building synthetic communities.”
- “Representativeness, that is, how well do these collections represent natural populations, is a key issue and is inherently difficult to achieve and determine. For estimation, cross-referencing against direct sequencing approaches from environmental samples is important. “
- “So far, strain collections have largely focused on bacteria; however, in order to be comprehensive, cultivation efforts need to be extended to include fungi, oomycetes, and, for some plants, archaea, to reflect the microbial diversity in planta.”
- “Extended culture collections will not only increase resemblance to the natural microbiota and facilitate synthetic microbiome research, but also represent a highly valuable resource in themselves“
- “As emphasized above, collections of microbial strains but also of their genomes will allow functional assessment.“
- “Genomic databases of culture collections enable the shift from phylogenetic to functional analysis, which is required to understand the plant-microbe interplay“
- Gnotobiotics
- To extend the biological toolbox of plants and associated microorganisms, gnotobiotic systems need further adaptations.
Ben O Oyserman, Marnix H Medema, Jos M Raaijmakers. Road MAPs to engineer host microbiomes. Current Opinion in Microbiology 2018, 43:46–54
- Emphasizes importance of microbiome associated phenotypes as a key tool
Sergaki et al. 2018. Challenges and Approaches in Microbiome Research: From Fundamental to Applied
- One key factor: “Efforts to Isolate, Characterize, and Use Microbial Strains in Synthetic Communities“
- Another key : “bridging the lab field gap – Limitations on the Experiments Performed in Controlled Conditions (The Lack of Context)”
Douglas AE (2018) Which experimental systems should we use for human microbiome science? PLoS Biol 16(3): e2005245. https://doi.org/10.1371/journal.pbio.2005245
- Some key quotes
- From abstract: “In this essay, I argue for thoughtful choice of model systems for human microbiome science. A greater variety of experimental systems, including wider use of invertebrate models, would benefit biomedical research, while systems ill-suited to experimental and genetic manipulation can be used to address very limited sets of scientific questions. Microbiome science benefits from the coordinated use of multiple systems, which is facilitated by networks of researchers with expertise in different experimental systems.”
- “Two linked factors play an important role in the appropriate choice of experimental systems for human microbiome research: history and purpose.”
- Extensive discussion of mouse as a model system, good and bad
- Other possible models: “What about the systems that are being used to study animal–microbe interactions but lack the in-depth infrastructure available to the traditional models? A number of systems used by relatively small communities of researchers have yielded major insights of general significance.”
- “One powerful solution is already starting to happen: networks of researchers who can share access to multiple different systems (traditional models, nontraditional models, in vitro systems, and human research) and the associated expertise. These networks can be (partly or entirely) colocated as centers at one (or several) institution(s)”
Animal Models for Microbiome Research: Advancing Basic and Translational Science: Proceedings of a Workshop (2018)
Chapter: 3 Non-Rodent Models for Microbiome Research
A. E. Douglas. Simple animal models for microbiome research. Nature Reviews Microbiology volume 17, pages 764–775(2019)
- recently published
Common principles and best practices for engineering microbiomes. Nature Reviews Microbiology volume 17, pages725–741(2019)
- Recently published.
- Tons of useful material in this but just published so have not taken extensive notes
- Abstract:
- We argue that structuring research and technology developments around a design–build–test–learn (DBTL) cycle will advance microbiome engineering and spur new discoveries of the basic scientific principles governing microbiome function.
- In this Review, we present key elements of an iterative DBTL cycle for microbiome engineering, focusing on generalizable approaches, including top-down and bottom-up design processes, synthetic and self-assembled construction methods, and emerging tools to analyse microbiome function.
Other papers focusing on why specific systems are good models
- Dissecting cause and effect in host-microbiome interactions using the combined worm-bug model system
- Zhang F, Berg M, Dierking K, Felix MA, Shapira M, Samuel BS, et al. Caenorhabditis elegans as a model for microbiome research. Front Microbiol. 2017;8:485.
- Trinder M, Daisley BA, Dube JS, Reid G. Drosophila melanogaster as a high-throughput model for host-microbiota interactions. Front Microbiol. 2017;8:751.
- Melancon E, De La Torre Canny S, Sichel S, Kelly M, Wiles TJ, Rawls JF, et al. Best practices for germ-free derivation and gnotobiotic zebrafish husbandry. Methods Cell Biol. 2017;138:61–100. Epub 2017/01/29.
- Macpherson AJ, McCoy KD. Standardised animal models of host microbial mutualism. Mucosal Immunol. 2015;8(3):476–86.
- McFall-Ngai M. Divining the essence of symbiosis: insights from the squid-vibrio model. PLoS Biol. 2014;12(2):e1001783.
- Fritz JV, Desai MS, Shah P, Schneider JG, Wilmes P. From meta-omics to causality: experimental models for human microbiome research. Microbiome. 2013;1(1):14.
Also see:
- Microbiome tractability and translation collection: https://www.nature.com/collections/bdfaigbece
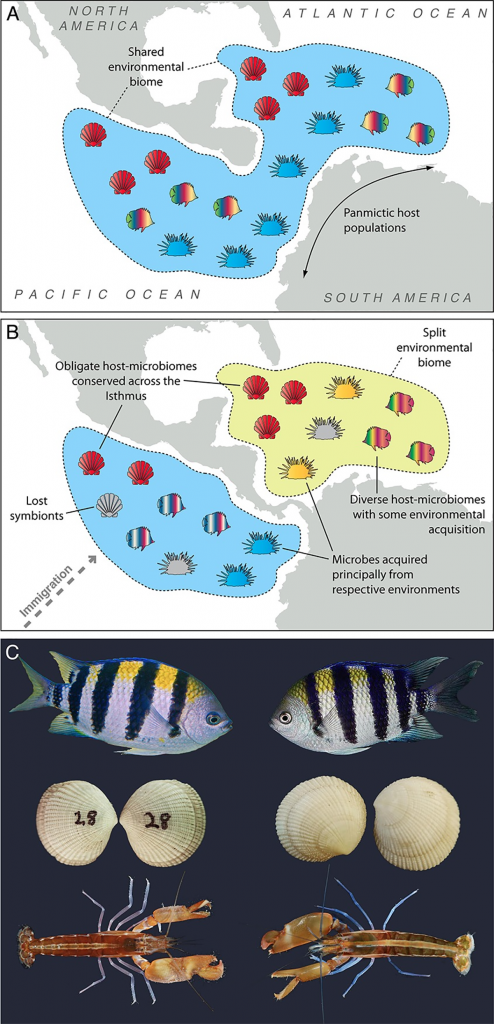
I think it would be helpful to clarify the objective here. Is it to study symbioses in general, or mutualisms in particular? If it’s the latter and the interest lies in the role of the symbionts, then I think a robust understanding of host biology is critical. Abudefduf (the fish in figure C above) are a case in point. They have very low levels of short-chain fatty acids in the gut. So although their gut microbiota must play some role in host health, hindgut fermentation will be trivial for host nutrition, unlike in other animals such as horses, pigs and humans.
Well, my goal here would be to develop model systems that would allow one to study all aspects of host-microbiome interactions. My focus personally would not be solely on mutualism.