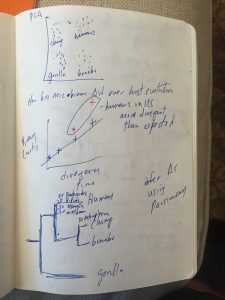
Andrew Moeller gave a talk at UC Davis Tuesday on “The evolution of the human gut microbiome”. He is a post doc at UC Berkeley working in the Nachman lab. I did not have a working computer so – gasp – I took notes with paper and pen. The talk was quite interesting and I thought …
The NY Times is reporting that an outbreak of Shigellosis in Flint may be connected to the water crisis there (in that people’s changes in their hand cleaning and other washing behaviors due to the water may have contributed to the outbreak). See the story (linked below) for more details. If true this is yet …
Just a mini post here. Got pointed to this press release of possible interest on a collaboration to study microbes on the space station and on other spacecrafts using single cell approaches: Source: NASA, Bigelow Laboratory Study Microbes on Spacecrafts – Bigelow Laboratory for Ocean Sciences
Worth checking this out I think. Source: Contributions of pioneering women in indoor environment and health – Nazaroff – 2016 – Indoor Air – Wiley Online Library From the introduction: On occasion, this journal has recounted historical achievements in the indoor air sciences. Sundell[1] provided a broad-ranging overview. I have written about the history of …
Definitely worth checking this out. Recent molecular studies have identified substantial fungal diversity in indoor environments. Fungi and fungal particles have been linked to a range of potentially unwanted effects in the built environment, including asthma, decay of building materials, and food spoilage. The study of the built mycobiome is hampered by a number of …
Interesting short new paper in arXiv: [1609.05763] Integrating citizen science with online learning to ask better questions Vineet Pandey, Scott Klemmer, Amnon Amir, Justine Debelius, Embriette R. Hyde, Tomasz Kosciolek, Rob Knight Abstract: Online learners spend millions of hours per year testing their new skills on assignments with known answers. This paper explores whether framing research questions …
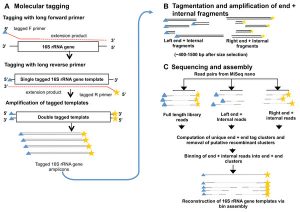
Definitely worth checking this one (from Burke and Darling) out if you do microbial diversity studies … Background The bacterial 16S rRNA gene has historically been used in defining bacterial taxonomy and phylogeny. However, there are currently no high-throughput methods to sequence full-length 16S rRNA genes present in a sample with precision. Results We describe …
New software/tool of possible interest: Source: MetaMLST: multi-locus strain-level bacterial typing from metagenomic samples Software: http://segatalab.cibio.unitn.it/tools/metamlst/ Abstract: Metagenomic characterization of microbial communities has the potential to become a tool to identify pathogens in human samples. However, software tools able to extract strain-level typing information from metagenomic data are needed. Low-throughput molecular typing schema such as Multilocus …
Collecting together here some reports from the Lake Arrowhead Microbial Genomes Meeting David Coil’s summaries Lake Arrowhead Microbial Genomics #LAMG16: Day 4 and Random Thoughts Lake Arrowhead Microbial Genomes #LAMG16: Day 3 Lake Arrowhead Microbial Genomics #LAMG16 Report: Day 2 Lake Arrowhead Microbial Genomics #LAMG16 Report: Day From me microbiology of the Built Environment session …
microBEnet sponsored a “Microbiology of the Built Environment” session at the Lake Arrowhead Microbial Genomes meeting earlier today. The schedule for the session is below Rachel Dutton University of California San Diego, La Jolla, CA “Genetics and Genomics of the Simple Microbial Communities from Cheese” Elizabeth Hénaff Weill Cornell Medicine, New York, NY “The Microbiome …