Genome-resolved metagenomics is the process of recovering de novo genomes belonging to individual organisms present in a community using DNA extracted from the whole community. These genomes are sometimes referred to as MAGs (metagenome-assembled genomes), and are recovered by associating assembled contigs together in “bins” in a process known as genome binning. As opposed to …
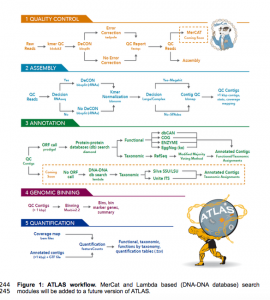
OK I have not tried this out yet but this could be of interest. ATLAS (Automatic Tool for Local Assembly Structures) is a comprehensive multi-omics data analysis pipeline that is massively parallel and scalable. ATLAS contains a modular analysis pipeline for assembly, annotation, quantification and genome binning of metagenomics and metatranscriptomics data and a framework …
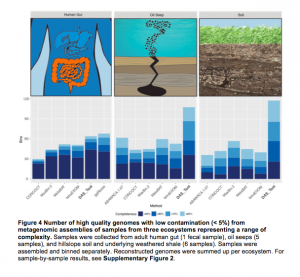
Saw an interesting Tweet check out this novel metagenomics tool to reconstruct thousands of high-quality genomes @ChrisMKSieber #banfieldlab https://t.co/67bk8CbFmP — Alexander Probst (@AlexJProbst) February 13, 2017 And this tool may be of interest – it is from a new preprint in BioRXiv. See abstract below: Microbial communities are critical to ecosystem function. A key objective …