Just wrote a post a few minutes ago about the sequencing and analysis of the genome(s) of representatives of the TM6 phylum of bacteria that were found in a hospital sink biofilm: First genome of TM6 — a novel phylum of bacteria — determined from a hospital sink sample.
And lo and behold just realized there is a second paper from the same group also dealing with a genome from an organism from a hospital sink biofilm: Genome of the pathogen Porphyromonas gingivalis recovered from a biofilm in a hospital sink using a high-throughput single-cell genomics platform. The new paper is conceptually similar to the first one but this time they focused on cells from the biofilm that were much more closely related to organisms that have been grown in the lab.
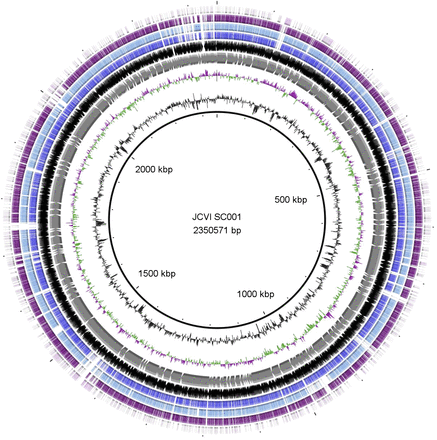
They obtain a nearly complete genome of a close relative of the pathogen Porphyromonas gingivalis using a combination of genome amplification, sequencing, and use of a new assembler.
And they use the genome to do some interesting comparative and evolutionary genomic analysis including SNP analysis of key genes:
Anyway – the paper is worth a look for those interested in single cell genomics and also microbiology of the built environment.

New at #microBEnet: Another genome from an uncultured microbe from a hospital sink biofilm #JCVI #Porphyromona… http://t.co/HCBhPbYjbl
RT @arturgreensward: New at #microBEnet: Another genome from an uncultured microbe from a hospital sink biofilm #JCVI #Porphyromona… http…
New at #microBEnet: Another genome from an uncultured microbe from a hospital sink biofilm #JCVI #Porphyromona… http://t.co/Z2DCF9T9Ni
RT @Dr_Bik: New at #microBEnet: Another genome from an uncultured microbe from a hospital sink biofilm #JCVI #Porphyromona… http://t.co/Z…
Some links to stories / press releases / press about this paper
http://www.jcvi.org/cms/press/press-releases/full-text/article/venter-institute-led-team-recovers-and-sequences-genome-of-periodontal-pathogen-from-biofilm-in-hosp/
http://www.genomeweb.com/blog/week-genome-research-65
http://www.myfoxwausau.com/story/21891598/venter-institute-led-team-recovers-and-sequences-genome-of-periodontal-pathogen-from-biofilm-in-hospital-sink-using-single-cell-genomics
http://www.utsandiego.com/news/2013/apr/09/tp-la-jolla-researchers-spot-single-microbe-cell/