OK I have not tried this out yet but this could be of interest.
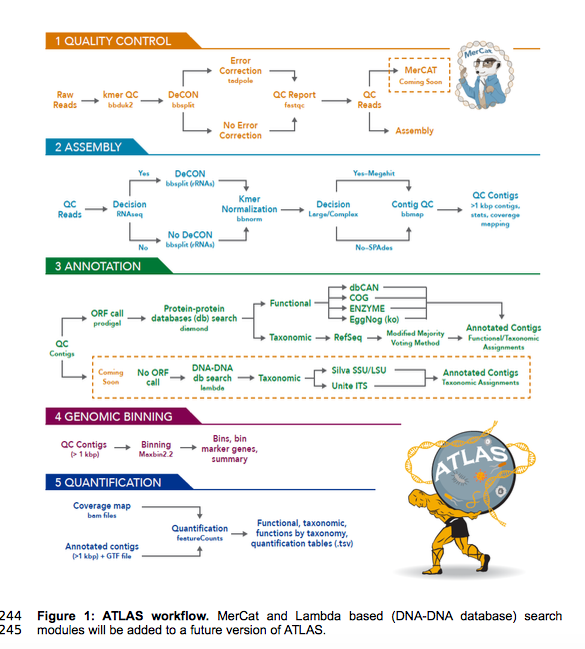
ATLAS (Automatic Tool for Local Assembly Structures) is a comprehensive multi-omics data analysis pipeline that is massively parallel and scalable. ATLAS contains a modular analysis pipeline for assembly, annotation, quantification and genome binning of metagenomics and metatranscriptomics data and a framework for reference metaproteomic database construction. ATLAS transforms raw sequence data into functional and taxonomic data at the microbial population level and provides genome-centric resolution through genome binning. ATLAS provides robust taxonomy based on majority voting of protein-coding open reading frames (ORFs) rolled-up at the contig level using modified lowest common ancestor (LCA) analysis. ATLAS is user-friendly, easy install through bioconda maintained as open-source on GitHub, and is implemented in Snakemake for modular customizable workflows.
Full citation:
White III RA, Brown J, Colby S, Overall CC, Lee J, Zucker J, Glaesemann KR, Jansson C, Jansson JK. (2017) ATLAS (Automatic Tool for Local Assembly Structures) – a comprehensive infrastructure for assembly, annotation, and genomic binning of metagenomic and metatranscriptomic data. PeerJ Preprints 5:e2843v1 https://doi.org/10.7287/peerj.preprints.2843v1
It is a preprint so it has not been peer reviewed formally yet. But the code is available at GitHub https://github.com/pnnl/atlas
Note – I first found out about this via Twitter
a fully automated pipeline for genome-resolved metagenomics. I wonder how it performs!https://t.co/sg4Rr8qfGi
— Alexander Probst (@AlexJProbst) March 5, 2017