So there is a new paper out that is incredibly interesting and has been getting lots of press coverage. The paper was in Nature: Endothelial TLR4 and the microbiome drive cerebral cavernous malformations. Sadly it is behind a paywall, so not everyone out there will have free access to it. But it is available in sci-hub for those who are interested in using that as a source (for more about sci-hub see here).
Basically, the paper is a very detailed analysis of the possible role the gut microbiome plays in cerebral cavernous malformations (CCMs). Here is their abstract:
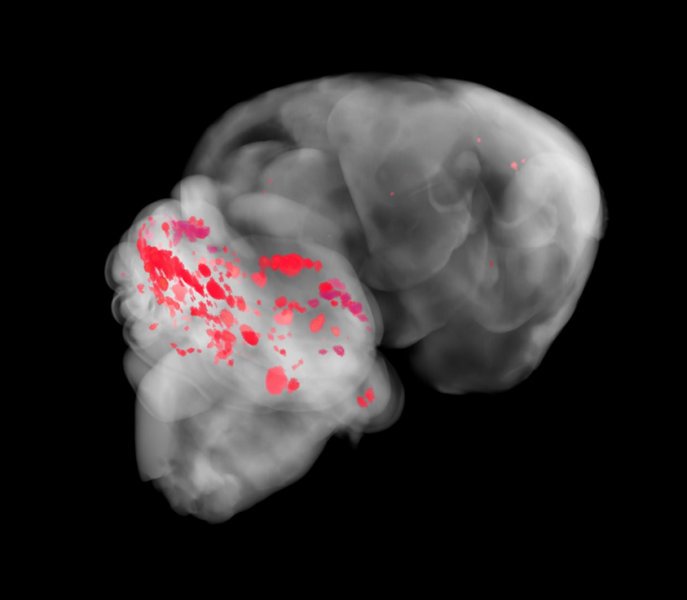
Cerebral cavernous malformations (CCMs) are a cause of stroke and seizure for which no effective medical therapies yet exist. CCMs arise from the loss of an adaptor complex that negatively regulates MEKK3–KLF2/4 signalling in brain endothelial cells, but upstream activators of this disease pathway have yet to be identified. Here we identify endothelial Toll-like receptor 4 (TLR4) and the gut microbiome as critical stimulants of CCM formation. Activation of TLR4 by Gram-negative bacteria or lipopolysaccharide accelerates CCM formation, and genetic or pharmacologic blockade of TLR4 signalling prevents CCM formation in mice. Polymorphisms that increase expression of the TLR4gene or the gene encoding its co-receptor CD14 are associated with higher CCM lesion burden in humans. Germ-free mice are protected from CCM formation, and a single course of antibiotics permanently alters CCM susceptibility in mice. These studies identify unexpected roles for the microbiome and innate immune signalling in the pathogenesis of a cerebrovascular disease, as well as strategies for its treatment.
And overall, I think the paper is quite nice. Some of the press about the paper is guilty of overselling their results (most of the key work is in mice, not humans, for example). But actually a lot of the press coverage has been pretty reasonable. A key thing about this study that I think is very important is that this is not really about the benefits that come from the microbiome. This is about the risks that come from the microbiome and possibly that come from some specific members of the community. And the authors argue that a possible way to limit health problems in people at risk for CCM is to treat them with antibiotics.
Anyway – I am not writing this to discuss CCM really. What I find fascinating is something pointed out to me by Paula Olsiewski from the Alfred P. Sloan Foundation. She sent me a link to a New York Times article about the study by Gina Kolata: A Baffling Brain Defect Is Linked to Gut Bacteria, Scientists Say – The New York Times.
This story has a really fascinating thread – quoted below with some highlighting by me.
A year ago, the scientists moved to a new building, and something unexpected happened. The experimental mice stopped developing the brain malformations.
Dr. Kahn’s student, Alan T. Tang, had been deleting the gene by injecting a drug into the abdomens of the mice. Sometimes a mouse would get an infection that would lead to an abscess, and bacteria leaked from the gut into the blood.
In the new building, only those mice still developed the brain defect. The other gene-deleted mice did not.
“When it happens three or four times, you realize this isn’t chance,” Dr. Kahn said.
He and his colleagues finally identified the culprit: Gram-negative bacteria, named for the way they stain, that carry a molecule in their cell walls, a lipopolysaccharide. Without a functioning gene, the lipopolysaccharide can signal veins in the brain to form blood bubbles.
In the old lab, mice without the gene carried enough Gram-negative bacteria to set off the disease. But in the new building, the mice were not exposed to the bacteria in sufficient amounts.
Really interesting in the context of “microbiology of the built environment” (aka MoBE) the field we are trying to cover here on microBEnet. I have not found any additional information about the move and exactly how they know that it was different microbes in the facility that contributed to this but I will keep digging around.
I have been suggesting for years that we need to monitor the built environment microbiome in the context of animal experiments such as this (e.g., would be good to have mouse facilities regularly collect data on the microbiome of the facility). And this certainly gives some support for that suggestion. I hope more people will start doing such MoBE characterization in the context of animal experiments to help document what, if any, role the microbes of the built environment play in findings.