Well, this paper is generating a lot of buzz on social media. Nucleic acid purification from plants, animals and microbes in under 30 seconds.
For example see Eric Topol’s Tweet about it
Now this is innovative: paper dipstick extraction of DNA or RNA in < 30 seconds https://t.co/RKuLsAW7fM @PLOSBiology @UQ_News pic.twitter.com/FH1krTFYMw
— Eric Topol (@EricTopol) November 21, 2017
Citation: Zou Y, Mason MG, Wang Y, Wee E, Turni C, Blackall PJ, et al. (2017) Nucleic acid purification from plants, animals and microbes in under 30 seconds. PLoS Biol15(11): e2003916. https://doi.org/10.1371/journal.pbio.2003916
Author summary: Nucleic acid amplification has proven to be indispensable in laboratories around the world for a myriad of applications from diagnostics to genotyping. The first step in any application aiming to amplify DNA or RNA is the extraction of nucleic acids from a complex biological sample; a task traditionally requiring specialised equipment, trained technicians, and multiple liquid handling steps. It is this complexity of current nucleic acid isolation methods that limit the use of many DNA amplification technologies outside of the modern laboratory environment. Therefore, in this study, we investigated new materials and approaches to simplify nucleic acid extraction. We found that cellulose-based filter paper can be used to rapidly bind nucleic acids, retain them during a short washing step to remove contaminants, and then elute them directly into the amplification reaction. We then adapted the cellulose filter to create a dipstick that can be used to purify nucleic acids from a wide range of plant, animal, and microbe samples in less than 30 seconds without the need for any specialised equipment. The speed and simplicity of our method makes it ideally suited for nucleic acid amplification-based applications both within and outside the laboratory, including limited resource settings such as remote field sites, developing countries, and teaching institutions.
Definitely seems interesting. However, it is unclear just how useful this will be for microbiome studies since they do not present anything about the effect of this protocol on microbial community analysis (e.g., maybe it only gets DNA from some taxa). Still – very interesting.
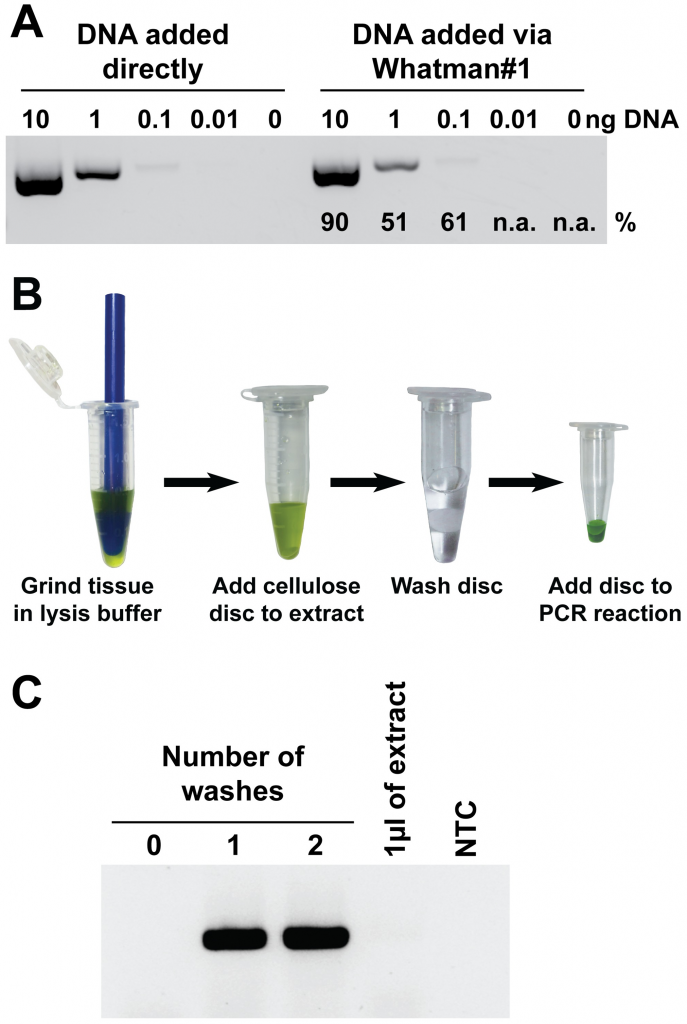
This paper is related:
Direct PCR Offers a Fast and Reliable Alternative to Conventional DNA Isolation Methods for Gut Microbiomes
Elin Videvall, Maria Strandh, Anel Engelbrecht, Schalk Cloete, Charlie K. Cornwallis
mSystems Nov 2017, 2 (6) e00132-17; DOI: 10.1128/mSystems.00132-17
It’s hard to tell if these methods are a step forward or backwards.
Increased cell lysis effort via physical or chemical disruption leads to a relatively higher recovery of gram positive and less representation of gram negatives, for example. Not a review here, but I think this is pretty consistent across many studies examining DNA extraction with NGS.
I have mostly used the MoBio methods due to widespread acceptance and Earth Microbiome Project Protocols, but these are costly methods (time and kit$). Anyone using the MoBio kits compared to direct extraction can appreciate the need to simplify methods. DNA extraction methods that require > pipetting and > elution steps will introduce > error The question is whether or not the benefits of additional purification steps outweigh errors associated with simpler methods.
Additionally, different DNA extraction methods offer access to different components of the microbiome by selectively extracting DNA. For example: https://doi.org/10.1016/j.mimet.2016.07.019