The last class! We started class with students taking the same survey that they took on the first day of class. This took about 30 minutes.
Then came time for students to analyze the Sanger sequences that they had prepared from the week before. Despite detailed instructions, students had a lot of questions in part due to the fact that every computer is a little bit different and/or this was their first time using the terminal. We were glad to have our entire teaching staff available to field questions and help interpret their chromatograms.
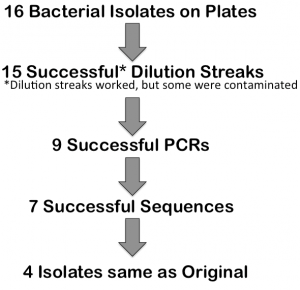
Once students had processed the Sanger sequences and filled in their results on the class data sheet, we had a discussion about what our overall results mean. We started with 16 isolates, and ended up with only 4 sequences that were identified to be the same as the original bug. Part of this could be chalked up to the scientific process, part of it was definitely due to a lack of sterile technique, and part of it was due to not using the most robust techniques (because of time and $). We discussed ways to continue the research, and what else we would do if we had the time and money to validate our results. So, did this CURE produce usable data? No, not really. Does this mean the CURE failed? Absolutely not! Students split into small groups and were assigned to one week of the CURE. Then as a class, we went through what we did each week and what transferable skills students were exposed to.

In summary students were exposed to: fundamentals of lab safety, accurate use of micropipette, oxidase and catalase tests, sterile technique, bacterial growth assays and subsequent analysis, gel electrophoresis, PCR clean up, DNA quantification, DNA dilutions, seqtrace software, BLAST search tool, Twitter for science communication, reading primary literature, and importance of open access publishing.