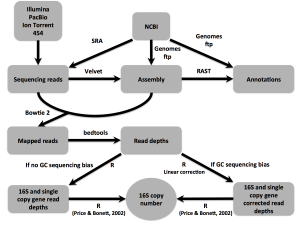
In scientific manuscripts, we tell stories of our research, generally in straight-line fashion with clear motivations and results. This type of research is rare (in my experience), with stories, motivations, and applications only realized post hoc. This is the nature of science, and our recent ISMEJ publication is no different. With “16Stimator: statistical estimation of …
When I first started trying to do PCR in Colleen Cavanaugh‘s lab in 1989, I was kind of on my own. Colleen was a newly hired profession at Harvard. She was busy getting things set up. And I was the only person in the lab – and I really knew very little. And basically I …
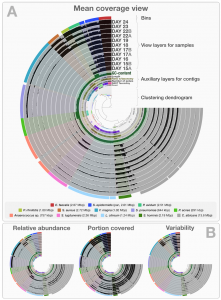
Three weeks ago I stood in front of the 60 attendees of the STAMPS course and asked, “How many of you are currently working with shotgun metagenomes?” Ten to fifteen people raised their hands. In contrast, almost all had their hands in the air when I asked how many were expecting to work with shotgun …
There is an interesting paper out a few days ago in PeerJ: MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. By Dongwan D. Kang, Jeff Froula, Rob Egan, Zhong Wang​. The key to what they do in the paper is summarized in Figure 1: The legend is below: There are three preprocessing steps before MetaBAT is …
About a year ago, I posted here about my free, interactive text: An Introduction to Applied Bioinformatics. The project is now Sloan-funded (as of April 2015), so there will be a lot of changes and expanded content coming down the pipe over the next few months. One major change that went in recently, and which …
There are few constants in this world. One exception, however, is the passing of day to night, which has gone on without fail since life first emerged on Earth. Early life quickly learned to anticipate changes associated with light and dark. This ability to tell time – to peer into the immediate future – was …
Just got this announcement by email and I thought it might be of interest: Join the curation effort & enroll in our first workshop! What: Workshop to produce curated 18S rDNA reference database. When: July 19th — 25th, 2015. Where: Vancouver, Canada on the University of British Columbia campus. How much: We will cover the cost of housing, breakfast and …
Dear metagenome method developers, The first challenge of the Initiative for the Critical Assessment of Metagenome Interpretation (CAMI) begins right now! Over the last three months, we received valuable feedback from the community playing with our toy data sets. We incorporated many of your suggestions, thanks again! Today, we proudly release the official data sets …
SciPy 2015 (Scientific Computing with Python) is coming up in Austin, TX this July 6-12. I attended SciPy last year for the first time to present on scikit-bio (see my talk here), and thought it was an excellent meeting. It was great to spend a week talking about software and software development, which isn’t the …
Have you ever wondered what’s in the air of a daycare center, where kids swap microbes every day? A team from Virginia Tech and the University of Pittsburgh is investigating the viral ecology in air of a daycare center using metagenomics. We have collected samples on HVAC filters and are looking for a sequencing core …