Do you think you can look at your microbiome or related data and detect patterns? Well, then this game may be for you: The 8-Bit Game That Makes Statistics Addictive Please make it stop. Nice article by Ed Yong in the Atlantic. But I warn you, don’t try it if you want to get …
I got an email the other day about a new feature at Figshare. It was about a new feature they have called “Collections“. But before I describe that I should probably describe Figshare. Figshare is a repository for depositing and sharing various digital objects including not just Figures (which they name kind of implies … …
I was casually browsing Twitter yesterday when something caught my eye DNA from dead #bacteria (and extruded DNA) may inflate species counts (depending on protocol). Early DNAse step? https://t.co/YQvUJq10Df – Michael J Wise (@some_wise) March 22, 2016 This Tweet embedded one from Carl Zimmer. Here is the original: The Earth is filled with relic DNA. https://t.co/eJ0lmggmAw …
So I had two thoughts when I read the article title “Indexed PCR Primers Induce Template-Specific Bias in Large-Scale DNA Sequencing Studies“… #1: Ack! Yet another thing to worry about in the insanely complicated craziness that is 16S amplicon sequencing. #2: I probably shouldn’t be the person to blog about this because I’ve honestly never …
In this new paper in MycoKeys we present a set of automatically updated lists of the most abundant fungal species hypotheses known from sequence data, but for which little to no taxonomic information is available. If anyone can shed light on the taxonomic affiliation of these species hypotheses, we’d be all ears. Through a “built environment” switch, …
The Marine Biological Laboratory in Woods Hole. MA will be offering their “STAMPS”: (Strategies & Techniques for Analyzing Microbial Population Structures) Course this August (8/3-8/13). Course Dates: August 3 — August 13, 2016 Deadline: April 8, 2016 | Apply here Course Website Course Schedule Directors: Mitchell L. Sogin, MBL and David B. Mark Welch, MBL …
Guest post from Trent Northen of LBNL. I wrote to Trent recently when I had seen a paper of his that I found fascinating: Exometabolite niche partitioning among sympatric soil bacteria. So I asked if he would be willing to write a guest post about it. And so – he did. It seems like the issues …
This paper by John Chase and crew posted recently on PeerJ takes a deep look at the office microbiome composition and concludes that the biggest factors affecting differences in swabbed samples are the geography of the office and where the sample was taken inside it. Their experiment showed minimal impact from other factors that are …
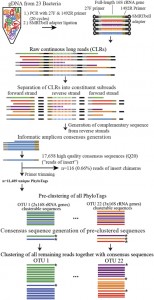
This seems like it could be of interest to microbial community researchers: The ISME Journal – High-resolution phylogenetic microbial community profiling Abstract (with bolding by me) Over the past decade, high-throughput short-read 16S rRNA gene amplicon sequencing has eclipsed clone-dependent long-read Sanger sequencing for microbial community profiling. The transition to new technologies has provided more …
From Pat Schloss: Hi mothur users, I wanted to let you know that I will be teaching another of my R workshops for microbial ecologists this May. The workshop will run from May 23rd to 25th near the Detroit airport. The workshop is being filled on a first come, first served basis. The workshop is …