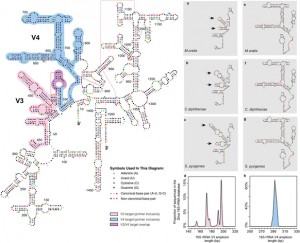
To date, characterization of ancient oral (dental calculus) and gut (coprolite) microbiota has been primarily accomplished through a metataxonomic approach involving targeted amplification of one or more variable regions in the 16S rRNA gene. For anyone interested in rRNA studies of microbial communities, ancient microbiomes, or analysis of samples with small amounts of DNA this …
Just got this from the Mothur email list: We continue to plug away at some really nice features to incorporate into mothur for your analytical pleasure. I wanted to let you know that the next mothur workshop will run from December 16 to 18 near the Detroit airport. The workshop is being filled on a …
As some QIIME users may be aware, we’re starting to think about our transition from QIIME 1 to QIIME 2. We want to briefly share our thoughts about this transition so the QIIME user community has an idea of what to expect as we start this process. See our post, Toward QIIME 2, on the QIIME …
Gearing up the UNITE database for the built mycobiome The team behind the UNITE database for molecular identification of fungi has been granted support from the Sloan Foundation to strengthen the support for fungi from the built environment. Launched in 2001 as an ITS database for identification of ectomycorrhizal fungi in the Nordic countries, UNITE …
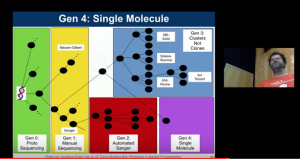
Every year for the last few years I have given a talk on the “Evolution of DNA Sequencing” at the “Workshop in Applied Phylogenetics” at Bodega Bay Marine Lab. I just did the talk and thought I would post the slides here. I note – I also added an evolutionary tree of sequencing methods which …
It is well known that antibiotic resistance in bacteria happens much faster than we can possibly develop novel antibiotics. So what if instead of trying to reinvent the wheel, we just rearrange it? Well, researchers at the H. Lee Moffitt Cancer Center and Research Institute had a similar idea in regards to reducing antibiotic resistance …
Norm Pace gave a talk at UC Davis yesterday on “Metagenomics and the Tree of Life”. I and a few other people posted live Tweets from the talk which I have compiled together via the Storify system. This “Storify” is embedded below. In addition, Lisa Cohen, who was at the talk posted her notes which …
Just got sent this by Katherine Bowman from the Board on Life Sciences. This is really important and if you know of someone who would be good please consider nominating them. Request for Committee Nominations — Microbiomes of the Built Environment: From Research to Application The National Academies of Sciences, Engineering, and Medicine are …
scikit-bio is a library implementing core bioinformatics algorithms and data structures, and providing support for bioinformatics data munging. You can learn more about the project by watching Jai Rideout and Evan Bolyen’s 20 minute SciPy 2015 talk. scikit-bio currently supports Python 2 (i.e., Legacy Python) and Python 3. We’re considering dropping support for Python 2, …
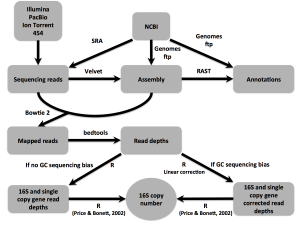
In scientific manuscripts, we tell stories of our research, generally in straight-line fashion with clear motivations and results. This type of research is rare (in my experience), with stories, motivations, and applications only realized post hoc. This is the nature of science, and our recent ISMEJ publication is no different. With “16Stimator: statistical estimation of …