Just came across this paper on bioRXiv. A Measure of Open Data: A Metric and Analysis of Reusable Data Practices in Biomedical Data Resources by Seth Carbon, Robin Champieux, Julie McMurry, Lilly Winfree, Letisha R Wyatt, and Melissa Haendel Abstract Data is the foundation of science, and there is an increasing focus on how data can be reused …
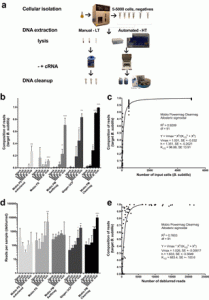
This may be of interest to people out there. Source: KatharoSeq Enables High-Throughput Microbiome Analysis from Low-Biomass Samples | mSystems Abstract: Microbiome analyses of low-biomass samples are challenging because of contamination and inefficiencies, leading many investigators to employ low-throughput methods with minimal controls. We developed a new automated protocol, KatharoSeq (from the Greek katharos [clean]), …
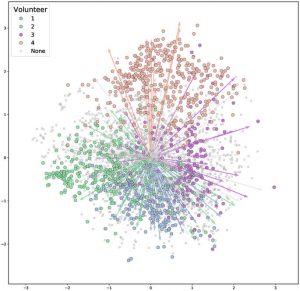
Article OK this new paper is pretty cool: Creating a 3D microbial and chemical snapshot of a human habitat | Scientific Reports. Abstract: One of the goals of forensic science is to identify individuals and their lifestyle by analyzing the trace signatures left behind in built environments. Here, microbiome and metabolomic methods were used to …
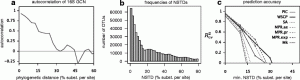
I entered the world of 16S rRNA gene metabarcoding as a naïve PhD student around five years ago. Like many people, I had hoped that there would have been others before me that had beaten the path and could guide me through the many pitfalls of this experimental approach. How wrong I was! I became …
rRNA gene copy number variation is still an issue in microbiome studies
The Gibbons group at the Institute for Systems Biology (ISB) is seeking a computationally-oriented Postdoctoral Fellow with interests in microbial ecology, evolution, and human health. The successful applicant will work with an interdisciplinary team of microbiologists, systems biologists, bioinformaticians, and clinical researchers to: Integrate and analyze multi-omic data from Arivale as part of the …
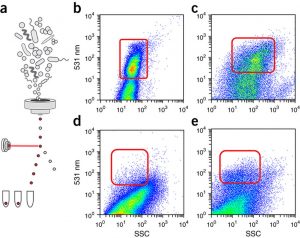
So you want to run FACS on your microbiome samples… By Cara Pardon Although optimized for eukaryotic cell research, an increasingly common technique used in the microbiome field is Fluorescence Activated Cell Sorting (FACS). From sorting cells for single cell genomics to identifying the host range of a plasmid; from detecting metabolically active cells in …
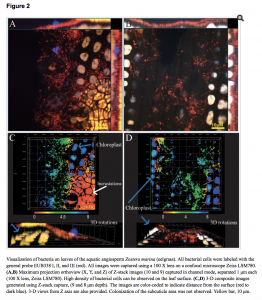
This paper may be of use to people working on plant microbiomes. Peredo EL and Simmons SL (2018) Leaf-FISH: Microscale Imaging of Bacterial Taxa on Phyllosphere. Front. Microbiol. 8:2669. doi: 10.3389/fmicb.2017.02669 Abstract Molecular methods for microbial community characterization have uncovered environmental and plant-associated factors shaping phyllosphere communities. Variables undetectable using bulk methods can play an …
This panel discussion’s topic is “Charting MoBE Research Priorities.” This was recorded at the MoBE 2017 symposium in Washington D.C. If you’re interested, check out all of the other speakers from MoBE 2017 on our YouTube channel!
I am hoping to find a system for everyone in my lab to use which would serve as lab notebooks for everyone and allow both written materials to be uploaded and also allow a mixture of different electronic materials such as bioinformatics workflows, pictures, videos, data files, etc. I have dug around a bit and …