New report on Synthetic Biology from the National Academies of Sciences, Engineering and Medicine that may be of interest. Synthetic biology expands the possibilities for creating new weapons — including making existing bacteria and viruses more harmful — while decreasing the time required to engineer such organisms, concludes a new report by the National Academies …
The Metcalf lab at Colorado State University is seeking a microbiome bioinformatics postdoctoral scientist. The scientist will analyze multi-omics data sets, including but not limited to: amplicon sequencing data, shotgun metagenomics data, and metabolomics data. Min. reqs. include: Ph.D. in bioinformatics, ecology, microbiology or similar field. Proficient in coding languages (Python preferred). Demonstrated knowledge of …
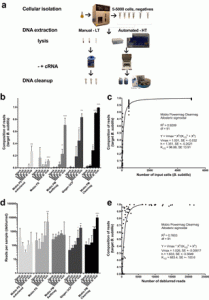
This may be of interest to people out there. Source: KatharoSeq Enables High-Throughput Microbiome Analysis from Low-Biomass Samples | mSystems Abstract: Microbiome analyses of low-biomass samples are challenging because of contamination and inefficiencies, leading many investigators to employ low-throughput methods with minimal controls. We developed a new automated protocol, KatharoSeq (from the Greek katharos [clean]), …
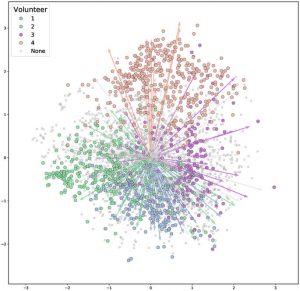
Article OK this new paper is pretty cool: Creating a 3D microbial and chemical snapshot of a human habitat | Scientific Reports. Abstract: One of the goals of forensic science is to identify individuals and their lifestyle by analyzing the trace signatures left behind in built environments. Here, microbiome and metabolomic methods were used to …
I have personally been interested in microbial forensics of various kinds for quite a while (I worked at TIGR for many years and was peripherally involved in some of the anthrax DNA sequencing and analysis there). I have even served on various working groups from the US Government discussing the potential for microbes to be …
Introduction In 2013-2014, a metagenomics project called “Pathomap” collected 1,457 swab samples from the surfaces of all active subway stations throughout New York City (NYC), as well as samples from the Gowanus Canal and several parks. Each sample was sequenced to an average depth of 3.6 million reads (paired-end 125 nucleotides), generating a city-wide metagenomic …