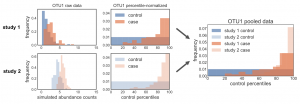
We recently published a method to normalize data that corrects for batch effects and allows for pooling of raw data across studies. In this post, I want to discuss more about the important (but potentially confusing) aspect of our method, where we add noise to the zeros before performing the normalization. I’ve gotten a few …
I just got pointed to this article today “Ugly Ducklings – the dark side of plastic materials in contact with potable water“, along with the accompanying commentary “About duck, pipes, and microbiomes“. I think this is a fascinating study for a few reasons. Firstly, like probably most parents, I observed and wondered about the biofilms …
We are a research group working on Genetics in a Vet School and closely collaborating with veterinary clinicians and dermatologists. We moved from specific pathogens, such as Leishmania, to specific commensals such as Demodex… and finally ended studying the whole: the skin microbiota on healthy dogs. Vets are always wondering why there are differences among …
So I had two thoughts when I read the article title “Indexed PCR Primers Induce Template-Specific Bias in Large-Scale DNA Sequencing Studies“… #1: Ack! Yet another thing to worry about in the insanely complicated craziness that is 16S amplicon sequencing. #2: I probably shouldn’t be the person to blog about this because I’ve honestly never …
A paper from this past year by GarcÃa-Mena et al entitled “Airborne Bacterial Diversity from the Low Atmosphere of Greater Mexico City” uses culture techniques and 16S sequencing of the air in Greater Mexico City. There doesn’t seem to be as much literature on microbes in the air as there is about other surfaces and parts of …
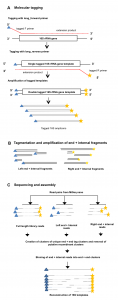
Microbial ecology has benefited enormously from the development of high throughput sequencing technologies, driving the discovery of massive diversity in environments from the ocean to the human body. Where sequencing of less than one hundred 16S rRNA genes from several samples used to be common place with cloning and Sanger sequencing, we can now generate tens …
Really nice article from Sarah Zhang at Gizmodo “Not Even Science Could Explain the Bacteria In My Apartment“. In it, she discusses the results of her participation in the Wildlife of Our Home study, the NY metagenomic data, and the fact that we’re still in the “exploration” phase of DNA-based microbial ecology. I think it’s …
Finally got around to reading “Impacts of Flood Damage on Airborne Bacteria and Fungi in Homes after the 2013 Colorado Front Range Flood” from the labs of Shelly Miller and Noah Fierer. The massive floods in 2013 provided the researchers with an opportunity to examine the lingering effects of flood damage, even post remediation. Ideally …
One of our goals at microBEnet is to increase communication and collaboration between people working in the disparate fields that make up the “microbiology of the built environment”. Most of what we’ve posted about in the past relates to general information and events… but we’d also like to host more technical information that would help …