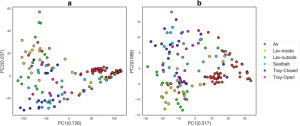
New paper of interest: The Airplane Cabin Microbiome | SpringerLink Abstract: Serving over three billion passengers annually, air travel serves as a conduit for infectious disease spread, including emerging infections and pandemics. Over two dozen cases of in-flight transmissions have been documented. To understand these risks, a characterization of the airplane cabin microbiome is necessary. …
rRNA gene copy number variation is still an issue in microbiome studies
This seems like it is a must read for anyone working on microbiomes: Frontiers | Microbiome Datasets Are Compositional: And This Is Not Optional | Microbiology Gloor GB, Macklaim JM, Pawlowsky-Glahn V and Egozcue JJ (2017) Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 8:2224. doi: 10.3389/fmicb.2017.02224 Summary from the paper: Datasets …
A while back I posted scans of some seminars from a while back here and I thought I would restart this activity (which I call retroblogging). I (and I hope some others) find it really interest to go back and see what people were talking about 10, 20, or more years ago. See for example Retroblogging …
Just a quick post here. There is a paper of interest I thought I would call attention to: Source: SILVA, RDP, Greengenes, NCBI and OTT — how do these taxonomies compare? | BMC Genomics by Monika Balvočiūtė and Daniel H. Huson Abstract: Background A key step in microbiome sequencing analysis is read assignment to taxonomic units. …
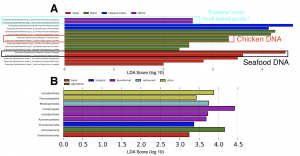
Amidst the November/December holiday chaos, myself and co-authors were proud to witness the publication of a neat new paper focused on ATM keypads in New York City. Yes, just like all other surfaces in the Built Environment, those ATM keypads are harboring lots of microbes and bits of orphaned DNA! This ATM keypad study was work that …
I was at a meeting a few weeks ago where the topic of fungal diversity surveys was discussed and many people there commented on how ITS based surveys (one of the main approaches for culture independent studies of fungi) had some limitations. ITS stands for the “internal transcribed sequence” and it is a region in …
In January 2014 I wrote this post about “microbiome” sequencing services. I have gotten many outside requests for the following information — what places (companies, Universities, government agencies, etc) provide contract services for rRNA PCR and sequencing? Source: Request — Information on Places that do rRNA sequencing as a service — microBEnet: the microbiology of the …
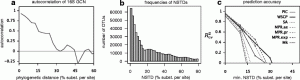
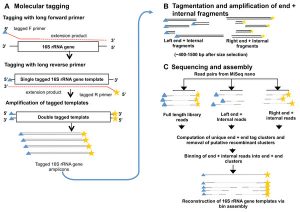
Definitely worth checking this one (from Burke and Darling) out if you do microbial diversity studies … Background The bacterial 16S rRNA gene has historically been used in defining bacterial taxonomy and phylogeny. However, there are currently no high-throughput methods to sequence full-length 16S rRNA genes present in a sample with precision. Results We describe …
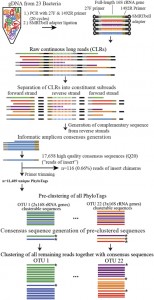
This seems like it could be of interest to microbial community researchers: The ISME Journal – High-resolution phylogenetic microbial community profiling Abstract (with bolding by me) Over the past decade, high-throughput short-read 16S rRNA gene amplicon sequencing has eclipsed clone-dependent long-read Sanger sequencing for microbial community profiling. The transition to new technologies has provided more …