(This blog post was prepared by students enrolled in the Koala Poop Microbiome Class in the Fall of 2016 at UC Davis)
The last seven weeks of laboratory and computer work have been a mixture of successes and failures that eventually brought us to Week 8. The goal of this week was to build phylogenetic trees for the bacteria identified last week. Last week we uploaded our BLAST results to RDP and referred back to those results to aid us in building phylogenetic trees.
But what exactly is a phylogenetic tree and why did we build some this week? A phylogenetic tree is a branched diagram that shows the evolutionary relationship between different species based on their physical and genetic characteristics (Reading Trees). The purpose of building phylogenetic trees is to further pinpoint what each of the bacteria we have been working with is by comparing it to an outgroup of the same family.
Here is a video that explains phylogenetic trees in more detail. https://www.youtube.com/watch?v=zXgSUdjsA_8
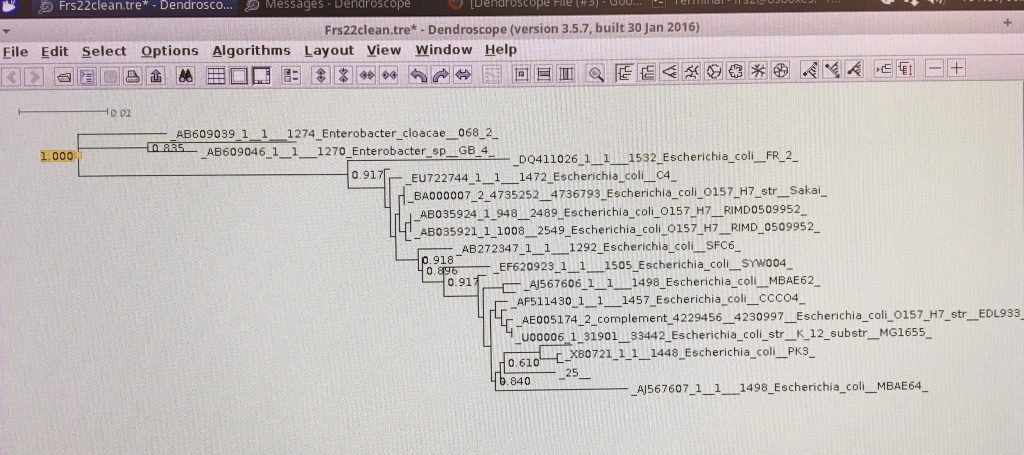
Building the phylogenetic trees was an interesting yet frustrating task since we were using unfamiliar programs to do something that most of the students had never done before. Most of the bacteria samples identified were Escherichia coli. We think that the number of species to choose from frustrated many students because it did not seem logistically possible to get the accurate trees we were working towards. As a result, some of the trees built were confusing to look at and appeared to be inaccurate. Another aspect of the work this week that some found to be frustrating was related to time-constraints; several students were not able to finish building their trees. Students were having to adjust to learning new technology in a short amount of time, but imagine having to do it all by hand! What we accomplished in a day would have taken exponentially longer without the use of computers!
Fun Fact:
Most of the programs utilized this week are designed to run for Unix/Linux operating systems. Since all the computers were running a windows operating system, in order to use the programs they were ran on a virtual machine.
Going forward, we are going to be testing antibiotic resistance/susceptibility on our identified bacteria. The identifying of our bacteria was extremely important because it will help us to make better predictions on how the antibiotics will affect the bacteria. If we know the bacteria, we know the structure of the bacteria: gram-negative, gram-positive, etc. This will do a great deal in determining which antibiotics are effective in treating these specific bacteria. It will also help to determine how the antibiotics interact with the tannins. This is crucial to the health of koalas.
Questions for the Science Community:
- What is a good genetic difference baseline for selecting an outgroup when building phylogenetic trees?
- What is a more efficient and practical method for selecting species that are generated from RDP?
Reference
“Reading Trees: A Quick Review.” Evolution.berkeley.edu. N.p., n.d. Web. 18 Nov. 2016.