The Gibbons group at the Institute for Systems Biology (ISB) is seeking a computationally-oriented Postdoctoral Fellow with interests in microbial ecology, evolution, and human health. The successful applicant will work with an interdisciplinary team of microbiologists, systems biologists, bioinformaticians, and clinical researchers to: Integrate and analyze multi-omic data from Arivale as part of the …
A post of interest from Noah Fierer. Let’s say a microbial ecologist wants to identify which bacteria are active in a given environmental sample at a given point in time. For example, you may want to determine which bacteria are active in a desert soil after a large rainfall event. This is often done …
I am getting ready for my “EVE161” class on Sequence based studies of the diversity of microbes. For more about past versions of the class see here: https://www.microbe.net/eve161/. Anyway – I am working on the reading for the class and I am looking for a good, recent, open access review of molecular phylogenetic methods. Any recommendations …
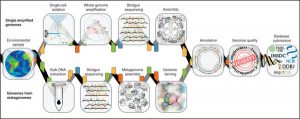
Genome-resolved metagenomics is the process of recovering de novo genomes belonging to individual organisms present in a community using DNA extracted from the whole community. These genomes are sometimes referred to as MAGs (metagenome-assembled genomes), and are recovered by associating assembled contigs together in “bins” in a process known as genome binning. As opposed to …
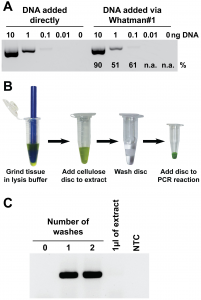
Well, this paper is generating a lot of buzz on social media. Nucleic acid purification from plants, animals and microbes in under 30 seconds. For example see Eric Topol’s Tweet about it Now this is innovative: paper dipstick extraction of DNA or RNA in < 30 seconds https://t.co/RKuLsAW7fM @PLOSBiology @UQ_News pic.twitter.com/FH1krTFYMw — Eric Topol (@EricTopol) …
For those interested in microbiology of the built environment this paper seems definitely worth checking out: Indoor microbiota in severely moisture damaged homes and the impact of interventions. Jayaprakash B, Adams RI, Kirjavainen P, Karvonen A, Vepsäläinen A, Valkonen M, Järvi K, Sulyok M, Pekkanen J, Hyvärinen A, Täubel M. Microbiome. 2017 Oct 13; 5(1):138 Abstract …
If you work on amplicon based microbial community analysis this is definitely worth a read. Source: A response to “Accuracy of microbial community diversity …” by R Edgar for QIIME users – Announcements – QIIME 2 Forum It is by Greg Caporaso in response to an article by Robert Edgar. I don’t want to get …
Of possible interest – a new paper (on which I am a coauthor) on standards for minimum information about single cell genomes and genomes assembled from metagenomes. See link below: Source: Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea : Nature Biotechnology : Nature Research
We are a research group working on Genetics in a Vet School and closely collaborating with veterinary clinicians and dermatologists. We moved from specific pathogens, such as Leishmania, to specific commensals such as Demodex… and finally ended studying the whole: the skin microbiota on healthy dogs. Vets are always wondering why there are differences among …
There is a wonderful tribute to Norm Pace in the Atlantic by Ed Yong. Norm Pace truly did blow the door off the microbial world. Source: Norm Pace Blew The Door Off The Microbial World – The Atlantic Lots of good stuff in there about Pace and his career. See for example this: Pace also …