A post of interest from Noah Fierer. Let’s say a microbial ecologist wants to identify which bacteria are active in a given environmental sample at a given point in time. For example, you may want to determine which bacteria are active in a desert soil after a large rainfall event. This is often done …
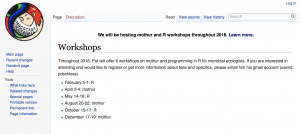
Just got this email and thought people might be interested: Hi mothur friends, Throughout 2018, I will offer 6 workshops on mothur and programming in R for microbial ecologists. Please see the Workshops page on the wiki for more information on what each workshop will cover. The dates are: February 5-7: R April 2-4: mothur …
I am getting ready for my “EVE161” class on Sequence based studies of the diversity of microbes. For more about past versions of the class see here: https://www.microbe.net/eve161/. Anyway – I am working on the reading for the class and I am looking for a good, recent, open access review of molecular phylogenetic methods. Any recommendations …
Genome-resolved metagenomics is the process of recovering de novo genomes belonging to individual organisms present in a community using DNA extracted from the whole community. These genomes are sometimes referred to as MAGs (metagenome-assembled genomes), and are recovered by associating assembled contigs together in “bins” in a process known as genome binning. As opposed to …
Thanks to rapid and relatively inexpensive techniques, anything that can be swabbed, scooped and sequenced is fair game for bacterial community analyses. And, microbiome-mania aside, we are still discovering incredible things about the invisible microbial world everyday. Fellow microBEnet author David Coil has suggested that it is timely to collate the wealth of information that …
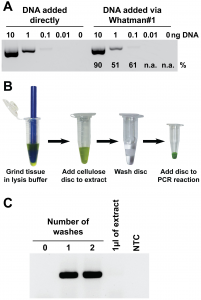
Well, this paper is generating a lot of buzz on social media. Nucleic acid purification from plants, animals and microbes in under 30 seconds. For example see Eric Topol’s Tweet about it Now this is innovative: paper dipstick extraction of DNA or RNA in < 30 seconds https://t.co/RKuLsAW7fM @PLOSBiology @UQ_News pic.twitter.com/FH1krTFYMw — Eric Topol (@EricTopol) …
This seems like it is a must read for anyone working on microbiomes: Frontiers | Microbiome Datasets Are Compositional: And This Is Not Optional | Microbiology Gloor GB, Macklaim JM, Pawlowsky-Glahn V and Egozcue JJ (2017) Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 8:2224. doi: 10.3389/fmicb.2017.02224 Summary from the paper: Datasets …
For those interested in microbiology of the built environment this paper seems definitely worth checking out: Indoor microbiota in severely moisture damaged homes and the impact of interventions. Jayaprakash B, Adams RI, Kirjavainen P, Karvonen A, Vepsäläinen A, Valkonen M, Järvi K, Sulyok M, Pekkanen J, Hyvärinen A, Täubel M. Microbiome. 2017 Oct 13; 5(1):138 Abstract …
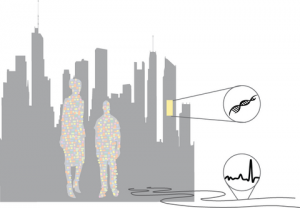
Hi MoBE blog, I just wanted to share a piece that I recently wrote for a Special Issue on the journal Microbial Biotechnology. The Special issue presents a collection articles about “The contribution of microbial biotechnology to sustainable development goals”, and the title of my article is “Microbial communities as biosensors for monitoring urban …
Collections of microbial cultures are critical tools in studying the microbial world around us. I have learned this from personal experience. The DSMZ for example, was absolutely essential to the entire GEBA (Genomic Encyclopedia of Bacteria and Archaea) project I coordinated via the DOE-JGI. I recommend that everyone out there find ways to support various …