In January 2014 I wrote this post about “microbiome” sequencing services. I have gotten many outside requests for the following information — what places (companies, Universities, government agencies, etc) provide contract services for rRNA PCR and sequencing? Source: Request — Information on Places that do rRNA sequencing as a service — microBEnet: the microbiology of the …
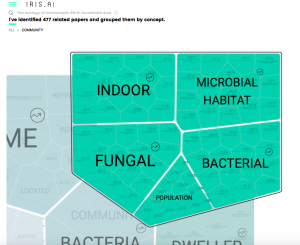
Got an email from the developers of IRIS AI the other day: And decided to play around with it. So I went to our Zotero collection on microbiology of the built environment: From there I got a paper and found it’s URL: Meadow, J. F., Altrichter, A. E., Kembel, S. W., Kline, J., Mhuireach, G., Moriyama, M., Northcutt, …
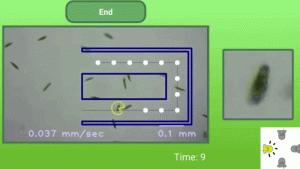
OMG. I want one. I want two. No – I want dozens. Caption from Gizmodo: “Introducing the LudusScope, a 3D-printed, open-sourced system that lets you control and play games with living microbes on your smartphone. Tormenting single-celled organisms has never been so much fun.” Source: This Smartphone Microscope Lets You Play Games With Microbes …
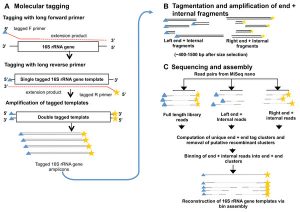
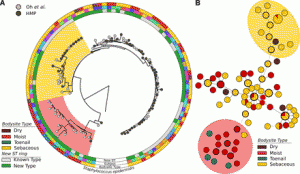
Definitely worth checking this one (from Burke and Darling) out if you do microbial diversity studies … Background The bacterial 16S rRNA gene has historically been used in defining bacterial taxonomy and phylogeny. However, there are currently no high-throughput methods to sequence full-length 16S rRNA genes present in a sample with precision. Results We describe …
New software/tool of possible interest: Source: MetaMLST: multi-locus strain-level bacterial typing from metagenomic samples Software: http://segatalab.cibio.unitn.it/tools/metamlst/ Abstract: Metagenomic characterization of microbial communities has the potential to become a tool to identify pathogens in human samples. However, software tools able to extract strain-level typing information from metagenomic data are needed. Low-throughput molecular typing schema such as Multilocus …
Ok so I made this into Clickbait. But you really should read this and that has nothing to do with me being a co-author. The paper is “Ten questions concerning the microbiomes of buildings” and it is in “Building and Environment” a journal that I am becoming more and more appreciative of every month. The …
Members of the QIIME development group, led by Greg Caporaso and Antonio Gonzalez, will teach a two-day workshop on bioinformatics tools for microbial ecology. The workshop will include lectures covering basic QIIME usage and theory, and hands-on work with QIIME to perform microbiome analysis from raw sequence data through publication-quality statistics and visualizations. The workshop …
Definitely worth checking this out. NIST is running a meeting on Standards for Microbiome Measurements From the site: This workshop will seek input on defining reference materials, reference data and reference methods for human microbiome community measurements. This workshop is sponsored by NIST and NIH’s National Institute of Allergy and Infectious Diseases and the Human Microbiome …
So I just saw this Tweet @BioMickWatson can’t find paper I’m thinking of, but OTU calling software/algorithm matters a lot – David Baltrus (@surt_lab) August 6, 2016 So I went to Mick Watson’s site and saw this: This is my attempt to collate the literature on how easy it is to introduce bias into microbiome …
On the heels of our 5000th citation, the QIIME 2 alpha release is now live and ready for testing! The best way to see where we are with QIIME 2 is to watch my SciPy 2016 presentation on QIIME 2 (the slides are available here). In this talk I give a bottom-up description of QIIME …