This is a pre-print that just came out from several UC Davis colleagues. I actually think the title is a bit misleading because these recommendations are a lot broader than just human fecal metagenomes. Definitely worth a read! Abstract below: Background Shotgun metagenomes are often assembled prior to annotation of genes which biases the functional …
Twitter Logo: Wikimedia Commons Public Domain Thought this might be of interest to various folks. I made a Twitter moment regarding a discussion about microbial genome annotation methods. Microbial Genome Annotation Methods https://platform.twitter.com/widgets.js
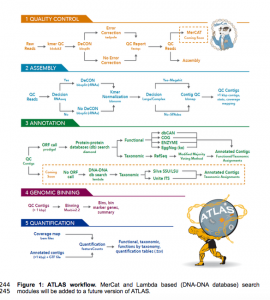
OK I have not tried this out yet but this could be of interest. ATLAS (Automatic Tool for Local Assembly Structures) is a comprehensive multi-omics data analysis pipeline that is massively parallel and scalable. ATLAS contains a modular analysis pipeline for assembly, annotation, quantification and genome binning of metagenomics and metatranscriptomics data and a framework …
We’d like to first thank Jon for the opportunity to discuss our work in this forum. We recently published a study investigating direct functional annotation of short metagenomic reads that stemmed from protocol development for our lab. Jon invited us to write a blog post on the subject, and we thought it would be a …
Posted a request for help on Twitter but I thought it might be of interest to some other people so am also posting about it here. Basically what we (me and Lizzy Wilbanks in my lab) is to take some microbial genomes and to fully annotate all of the repeat elements and all of the …