Just got this from the Mothur email list: We continue to plug away at some really nice features to incorporate into mothur for your analytical pleasure. I wanted to let you know that the next mothur workshop will run from December 16 to 18 near the Detroit airport. The workshop is being filled on a …
Norm Pace gave a talk at UC Davis yesterday on “Metagenomics and the Tree of Life”. I and a few other people posted live Tweets from the talk which I have compiled together via the Storify system. This “Storify” is embedded below. In addition, Lisa Cohen, who was at the talk posted her notes which …
Special Seminar Dr. Norman Pace Distinguished Professor Department of Molecular, Cellular and Developmental Biology University of Colorado, Boulder Metagenomics and the Tree of Life Tuesday, October 6th, 2015 1.30 pm Genome and Biomedical Science Facility (GBSF) 1005 Host: Jonathan Eisen (jaeisen@ucdavis.edu) For more about Pace’s work see http://pacelab.colorado.edu
Microbial ecology has benefited enormously from the development of high throughput sequencing technologies, driving the discovery of massive diversity in environments from the ocean to the human body. Where sequencing of less than one hundred 16S rRNA genes from several samples used to be common place with cloning and Sanger sequencing, we can now generate tens …
Another paper on how sample processing (and in this case PCR primer choice) can influence microbiome studies. And another one that is definitely worth looking at: 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 2015, 3:26 doi:10.1186/s40168-015-0087-4 by Alan W. Walker, Jennifer C. Martin, …
Continuing on my theme of scanning in notes from old seminars of interest. Here is one of definite interest to the microbial diversity crowd. These are notes from a talk by Norm Pace on February 7, 1990. My entire career was massively affected by interactions (mostly indirect) with Pace when I was an undergrad …
I got this email a few days ago from Jed Fuhrman. He had sent it to a group of people working on mcirobial diversity and he encouraged people to share it. I asked and he approved posting it here and I thought it would be of interest. (The image above is from Lane et al. 1985 …
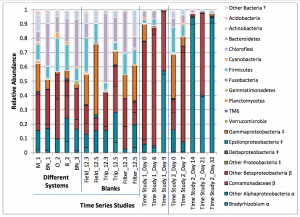
There is an interesting and potentially important new paper out from Caitlin Proctor, Marc Edwards and Amy Pruden: Microbial composition of purified waters and implications for regrowth control in municipal water systems in Environmental Science: Water Research & Technology. The abstract is below:
Really nice collection in the journal RNA for their 20th anniversary. So many interesting things here on the RNA world, RNA ctalysis, RNA structure, RNA function and more. Although all of these papers in some way relate to the work I do on sequencing and analyzing microbial genomes and metagenomes, a few of of particular …
So many new tools and methods in microbiome and microbial community studies and it is just really hard to keep up with them. Here are some that have caught my eye recently: PLOS ONE: IM-TORNADO: A Tool for Comparison of 16S Reads from Paired-End Libraries. Jeraldo P, Kalari K, Chen X, Bhavsar J, Mangalam A, …