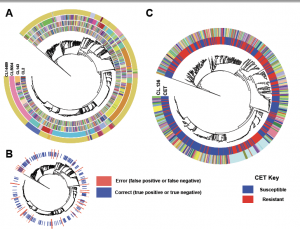
This preprint seems like it may be of interest to folks: Precise prediction of antibiotic resistance in Escherichia coli from full genome sequences | bioRxiv Basically, the authors showed that, using a machine learning approach, they can quite accurately predict antibiotic resistance in E. coli from whole genome data. The emergence of microbial antibiotic resistance …
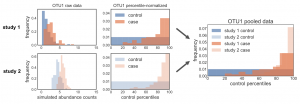
We recently published a method to normalize data that corrects for batch effects and allows for pooling of raw data across studies. In this post, I want to discuss more about the important (but potentially confusing) aspect of our method, where we add noise to the zeros before performing the normalization. I’ve gotten a few …
This is really fascinating: Source: Team discover how microbes survive clean rooms and contaminate spacecraft Hat tip to Elisabeth Bik for posting about this on Twitter So cool: microbes actually feed on the cleaning products. Also cool: undergraduates as authors. Team discover how microbes survive clean rooms and contaminate spacecraft https://t.co/ox4kMawHko — Elisabeth Bik (@MicrobiomDigest) June …
So we recently had two problems with a common solution: #1, we hadn’t had a lab social function in awhile #2, we wanted to try using the MinION sequencer in a “field” setting, but weren’t quite ready to try it without electricity on hand. The obvious solution, have the entire lab over to my house …
The Metcalf lab at Colorado State University is seeking a microbiome bioinformatics postdoctoral scientist. The scientist will analyze multi-omics data sets, including but not limited to: amplicon sequencing data, shotgun metagenomics data, and metabolomics data. Min. reqs. include: Ph.D. in bioinformatics, ecology, microbiology or similar field. Proficient in coding languages (Python preferred). Demonstrated knowledge of …
Hotel pools and spas cause a third of pool-linked disease outbreaks Just a quick one here — From NBC: Here’s what’s in your pool this summer
Interesting new article from Rob Dunn’s lab about the microbial and arthopod communities associated with chimpanzee beds. The article itself is clearly pitched within a microbiology of the built environment context… highlighting the fact that we haven’t really looked much a non-human constructed artificial environments. Pretty cool stuff! The outpouring of press on the topic …
So – just a quick post for now. More details to come. But I wanted to get a little bit about this out there. For a few years I have been teaching a mid-level course at UC Davis on “DNA sequencing based studies of microbial diversity’. The course has evolved over the years from a …
This should be of interest to many: mSystems has a whole issue dedicated to essays by “Early-Career Systems Microbiology Scientists”. Early-Career Systems Microbiology Scientists Jack A. Gilbert mSystems March/April 2018 3:e00002-18; doi:10.1128/mSystems.00002-18 http://msystems.asm.org/content/3/2/e00002-18 Conflict of Interest Declarations by Contributing Editors of the Special Issue on Early-Career Systems Microbiology Scientists, Sponsored by Janssen Human Microbiome …
(This is a blog post by Cinta Gomez-Silvan, the first author on this paper) Story behind the paper: A comparison of methods used to unveil the genetic and metabolic pool in the built environment. It seems pretty obvious that in the field of microbial ecology we need to understand how living microorganism interact. Nevertheless, …