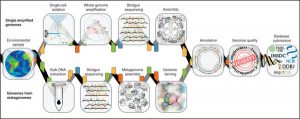
Genome-resolved metagenomics is the process of recovering de novo genomes belonging to individual organisms present in a community using DNA extracted from the whole community. These genomes are sometimes referred to as MAGs (metagenome-assembled genomes), and are recovered by associating assembled contigs together in “bins” in a process known as genome binning. As opposed to …
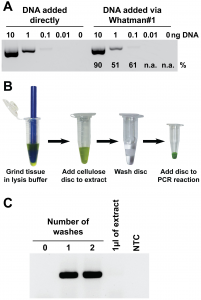
Well, this paper is generating a lot of buzz on social media. Nucleic acid purification from plants, animals and microbes in under 30 seconds. For example see Eric Topol’s Tweet about it Now this is innovative: paper dipstick extraction of DNA or RNA in < 30 seconds https://t.co/RKuLsAW7fM @PLOSBiology @UQ_News pic.twitter.com/FH1krTFYMw — Eric Topol (@EricTopol) …
This seems like it is a must read for anyone working on microbiomes: Frontiers | Microbiome Datasets Are Compositional: And This Is Not Optional | Microbiology Gloor GB, Macklaim JM, Pawlowsky-Glahn V and Egozcue JJ (2017) Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 8:2224. doi: 10.3389/fmicb.2017.02224 Summary from the paper: Datasets …
Hi MoBE blog, I just wanted to share a piece that I recently wrote for a Special Issue on the journal Microbial Biotechnology. The Special issue presents a collection articles about “The contribution of microbial biotechnology to sustainable development goals”, and the title of my article is “Microbial communities as biosensors for monitoring urban …
If you work on amplicon based microbial community analysis this is definitely worth a read. Source: A response to “Accuracy of microbial community diversity …” by R Edgar for QIIME users – Announcements – QIIME 2 Forum It is by Greg Caporaso in response to an article by Robert Edgar. I don’t want to get …
This new paper in PLOS Computational. Biology may be of interest: : The application of project-based learning in bioinformatics training by Emery LR and Morgan SL. Although many out there may already be doing projects based learning for bioinformatics, many others are not. This paper discusses how the authors added a projects component to an existing course …
Of possible interest – a new paper (on which I am a coauthor) on standards for minimum information about single cell genomes and genomes assembled from metagenomes. See link below: Source: Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea : Nature Biotechnology : Nature Research
We are a research group working on Genetics in a Vet School and closely collaborating with veterinary clinicians and dermatologists. We moved from specific pathogens, such as Leishmania, to specific commensals such as Demodex… and finally ended studying the whole: the skin microbiota on healthy dogs. Vets are always wondering why there are differences among …
There is a wonderful tribute to Norm Pace in the Atlantic by Ed Yong. Norm Pace truly did blow the door off the microbial world. Source: Norm Pace Blew The Door Off The Microbial World – The Atlantic Lots of good stuff in there about Pace and his career. See for example this: Pace also …
Just saw this story and thought people might be interested in it. Source: Brain-Invading Tapeworm That Eluded Doctors Spotted by New DNA Test – Scientific American The article discusses a new test to be offered by UCSF as well as some other tests from other places that all involve some type of metagenomic sequencing to …