The Metcalf lab at Colorado State University is seeking a microbiome bioinformatics postdoctoral scientist. The scientist will analyze multi-omics data sets, including but not limited to: amplicon sequencing data, shotgun metagenomics data, and metabolomics data. Min. reqs. include: Ph.D. in bioinformatics, ecology, microbiology or similar field. Proficient in coding languages (Python preferred). Demonstrated knowledge of …
So – just a quick post for now. More details to come. But I wanted to get a little bit about this out there. For a few years I have been teaching a mid-level course at UC Davis on “DNA sequencing based studies of microbial diversity’. The course has evolved over the years from a …
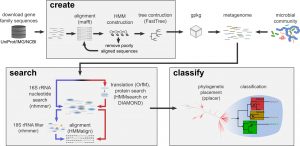
This could be of use to many people: Source: GraftM: a tool for scalable, phylogenetically informed classification of genes within metagenomes | Nucleic Acids Research | Oxford Academic Large-scale metagenomic datasets enable the recovery of hundreds of population genomes from environmental samples. However, these genomes do not typically represent the full diversity of complex microbial …
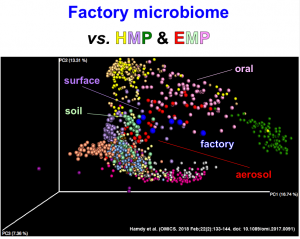
The story behind: Toward the Drug Factory Microbiome: Microbial Community Variations in Antibiotic-Producing Clean Rooms (OMICS. 2018 Feb;22(2):133-144. doi: 10.1089/omi.2017.0091.) PMID: 28873001 [Preprint] It was in 2013, on my return to Cairo University after a couple of years as a visiting scientist at UCSD, that I met Amal for the first time. Amal was a fresh master’s …
(Note: This post was updated on 3/26/18 after re-running some of our analysis based on suggestions in the comments and on Twitter). We were very excited to get our MinION sequencer from Nanopore 1.5 years ago… so many potential applications; sequencing in real time, use in the classroom, use in the field, not having to …
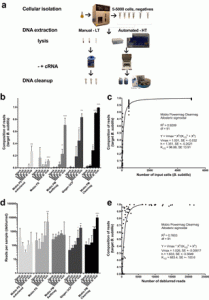
This may be of interest to people out there. Source: KatharoSeq Enables High-Throughput Microbiome Analysis from Low-Biomass Samples | mSystems Abstract: Microbiome analyses of low-biomass samples are challenging because of contamination and inefficiencies, leading many investigators to employ low-throughput methods with minimal controls. We developed a new automated protocol, KatharoSeq (from the Greek katharos [clean]), …
I entered the world of 16S rRNA gene metabarcoding as a naïve PhD student around five years ago. Like many people, I had hoped that there would have been others before me that had beaten the path and could guide me through the many pitfalls of this experimental approach. How wrong I was! I became …
The Gibbons group at the Institute for Systems Biology (ISB) is seeking a computationally-oriented Postdoctoral Fellow with interests in microbial ecology, evolution, and human health. The successful applicant will work with an interdisciplinary team of microbiologists, systems biologists, bioinformaticians, and clinical researchers to: Integrate and analyze multi-omic data from Arivale as part of the …
Just got this email and thought people might be interested: Hi mothur friends, Throughout 2018, I will offer 6 workshops on mothur and programming in R for microbial ecologists. Please see the Workshops page on the wiki for more information on what each workshop will cover. The dates are: February 5-7: R April 2-4: mothur …
I am getting ready for my “EVE161” class on Sequence based studies of the diversity of microbes. For more about past versions of the class see here: https://www.microbe.net/eve161/. Anyway – I am working on the reading for the class and I am looking for a good, recent, open access review of molecular phylogenetic methods. Any recommendations …