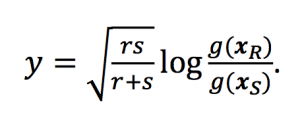Why use Bracken instead of Kraken? by Jennifer Lu and Steven Salzberg Kraken is a very fast, accurate program for classifying metagenomic sequencing data. It takes a set of reads, contigs, or other DNA sequences and for assigns a taxonomic label (species, genus, etc.) to each one. Last year we discovered that some people were …
WTH is ILR? by Alex Washburne & Jamie Morton Jamie and I have penned a popularization of the isometric log-ratio transform with the intention to allow non-mathematicians to understand the intuition behind what it is, why we use it, and how different methods use the same tool in different ways. The full write-up is available …
I thought this might be of interest. [View the story “Interesting discussion of data vs. metadata started at #NeLLi17 & continuing” on Storify]
Members of the QIIME development group, led by Greg Caporaso, will teach a three-day workshop on bioinformatics tools for microbial ecology. The workshop will include lectures covering basic QIIME 2 usage and theory, and hands-on work with QIIME 2 to perform microbiome analysis from raw sequence data through publication-quality statistics and visualizations. This workshop will …
Just a quick post here. There is a paper of interest I thought I would call attention to: Source: SILVA, RDP, Greengenes, NCBI and OTT — how do these taxonomies compare? | BMC Genomics by Monika Balvočiūtė and Daniel H. Huson Abstract: Background A key step in microbiome sequencing analysis is read assignment to taxonomic units. …
I got an email from a colleague requesting some help on an informatics issue and I thought it might be useful to post it here. I have been thinking about starting a section of this blog on “Technical Help Requests” or something like that so I guess this is a test. Here is the request …
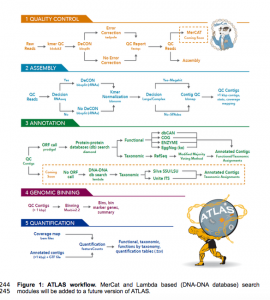
OK I have not tried this out yet but this could be of interest. ATLAS (Automatic Tool for Local Assembly Structures) is a comprehensive multi-omics data analysis pipeline that is massively parallel and scalable. ATLAS contains a modular analysis pipeline for assembly, annotation, quantification and genome binning of metagenomics and metatranscriptomics data and a framework …
I have personally been interested in microbial forensics of various kinds for quite a while (I worked at TIGR for many years and was peripherally involved in some of the anthrax DNA sequencing and analysis there). I have even served on various working groups from the US Government discussing the potential for microbes to be …
Hi All I’d just like to highlight a job we are advertising, to lead the bioinformatic (sequence+variation) analysis of 100,000 M tuberculosis genomes which we are sequencing (WGS), ~50,000 of which will be phenotyped for 12 drugs, and the remainder for some subset. Note the deadline is very soon – Monday 6th March! This project …
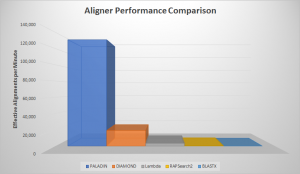
With our paper recently accepted for publication and many months of practical application, we’re excited to spread the word about our new tool for characterizing metagenomes, PALADIN! Short for Protein ALignment And Detection INterface, PALADIN was our response to the inherent problem of limited references in shotgun metagenomic sequencing. While DNA barcoding involving 16S, 18S, …