We just published an extended genome report from eight bacteria that were sequenced as part of the “Built Environment Reference Genome” (BERG) project. BERG is a microBEnet run effort to increase the number of reference genomes in the Built Environment (BE) by offering free genome sequencing to a number of labs working in the field. …
Quick post here sharing an interesting sounding event “2017 Innovation Lab on Quantitative Approaches to Biomedical Data Science Challenges in our Understanding of the Microbiome“. From the website: The BD2K Training Coordinating Center is organizing an Innovation Lab to foster new interdisciplinary collaborations among quantitative and biomedical researchers to address data science challenges in …
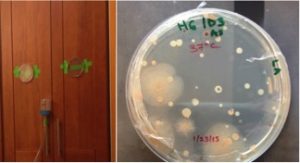
Here is a great chance to participate in a microbiology of the built environment citizen science project on gutters. “Examining this water…I found floating therein divers earthy particles, and some green streaks, spirally wound serpent-wise…and I judge that some of these little creatures were above a thousand times smaller than the smallest ones I have ever …
This is a great idea — Instructor training for teaching computational workshops. From my UC Davis colleague Titus Brown. I have copied details below. As part of our Summer Institute in Data Intensive Biology, we will be running a week-long instructor training from June 18 to June 25 at the University of California, Davis. The instructor training will …
Call for Papers: MoBE 2017 Special Issue of BioMed Central’s Microbiome Journal (Submission Guidelines) We invite submissions of MoBE papers highlighting recent research and emerging hot topics along the theme of “MoBE Research to Applications” for our peer-reviewed MoBE special issue. Publishing charges are sponsored by the MoBE meeting and BioMed Central’s Microbiome Journal. This special issue will be available by October 1st, …
I have been tracking #actuallivingscientist postings on Twitter compiled those by people doing microbiology. Actual living microbiologists. [View the story “These are actual living microbiologists #actuallivingscientist #microbiology #microbes #microBEnet” on Storify]
There is an increasing number of studies with a large number genomes recovered from isolate, metagenome, or single cell sequencing. To bridge the gap between the available genome sequences and available phenotype information, we have developed Traitar, a bioinformatics software to phenotype bacteria based on their genome sequence (see workflow below) . Traitar includes phenotype models for …
Of interest to many microBEnet readers. From the website: On behalf of the Organizing Committee, we cordially invite you to attend the Healthy Buildings 2017-Asia (HB2017-Asia), being held from September 2 to 5, 2017, at the College of Medicine, National Cheng Kung University in Tainan, Taiwan. The HB2017-Asia inscribes itself on the path of a …
We are happy to announce that registration for GSC19 is now open !! Theme: Extending Standards to Viruses and Microbial Eukaryotes Date: May 15th-17th, 2017 Location: Stamford Plaza, Brisbane, Australia Host: Philip Hugenholtz Australian Centre for Ecogenomics, University of Queensland The agenda is packed with exciting sessions and topics: Viral classification and …
There has been some interest in our recent preprint describing Oxford Nanopore MinIONTM sequencing for 16S rRNA microbiome characterization and I was asked to write a post for microbenet on this technology. Disclaimers – this paper is a work in progress – our paper has not yet been peer-reviewed and we are continuing to revise …