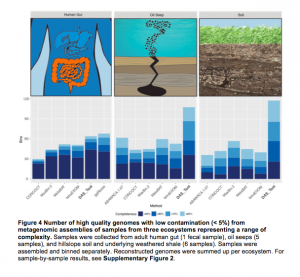
Saw an interesting Tweet check out this novel metagenomics tool to reconstruct thousands of high-quality genomes @ChrisMKSieber #banfieldlab https://t.co/67bk8CbFmP — Alexander Probst (@AlexJProbst) February 13, 2017 And this tool may be of interest – it is from a new preprint in BioRXiv. See abstract below: Microbial communities are critical to ecosystem function. A key objective …
There are a number of cases where determining the relationship between microbes is at the center of a research question. Are the microbes inhabiting a building the same as those inhabiting its tenants? Are the microbes in a hospital room the same as those that colonize newborn babies? Is the E. coli living on a …
There is an increasing number of studies with a large number genomes recovered from isolate, metagenome, or single cell sequencing. To bridge the gap between the available genome sequences and available phenotype information, we have developed Traitar, a bioinformatics software to phenotype bacteria based on their genome sequence (see workflow below) . Traitar includes phenotype models for …
There has been some interest in our recent preprint describing Oxford Nanopore MinIONTM sequencing for 16S rRNA microbiome characterization and I was asked to write a post for microbenet on this technology. Disclaimers – this paper is a work in progress – our paper has not yet been peer-reviewed and we are continuing to revise …
Amidst the November/December holiday chaos, myself and co-authors were proud to witness the publication of a neat new paper focused on ATM keypads in New York City. Yes, just like all other surfaces in the Built Environment, those ATM keypads are harboring lots of microbes and bits of orphaned DNA! This ATM keypad study was work that …
Kevin Van Den Wymelenberg and Jessica Green, of the Biology and the Built Environment Center (BioBE), are currently seeking a microbial ecology Research Associate / Research Assistant Professor / Research Associate Professor (non-tenure track faculty) to investigate fundamental questions surrounding the role of microorganisms (bacteria, archaea, fungi, protists, and viruses) in the built environment and …
The Department of Statistics at Oregon State University is hosting a summer REU (Research Experiences for Undergraduates) program that focuses on microbiome data analysis. This is a unique, paid research and training opportunity for STEM undergraduate students who want an interdisciplinary research experience in both biology and statistics. During a ten week training period, students will be …
Members of the QIIME development group, led by Greg Caporaso and Antonio Gonzalez, will teach a two-day workshop on bioinformatics tools for microbial ecology. The workshop will include lectures covering basic QIIME usage and theory, and hands-on work with QIIME to perform microbiome analysis from raw sequence data through publication-quality statistics and visualizations. The workshop …
On the heels of our 5000th citation, the QIIME 2 alpha release is now live and ready for testing! The best way to see where we are with QIIME 2 is to watch my SciPy 2016 presentation on QIIME 2 (the slides are available here). In this talk I give a bottom-up description of QIIME …
A few weeks ago we wrapped up Spring Quarter here at UC Davis and the end of our experimental “Swabs to Genomes” class, taught as a freshman seminar. As we introduced here, the idea was to take a set of students from colonies on a plate (from a swab) through a collection of ready to …