Microbial ecology has benefited enormously from the development of high throughput sequencing technologies, driving the discovery of massive diversity in environments from the ocean to the human body. Where sequencing of less than one hundred 16S rRNA genes from several samples used to be common place with cloning and Sanger sequencing, we can now generate tens …
When I first started trying to do PCR in Colleen Cavanaugh‘s lab in 1989, I was kind of on my own. Colleen was a newly hired profession at Harvard. She was busy getting things set up. And I was the only person in the lab – and I really knew very little. And basically I …
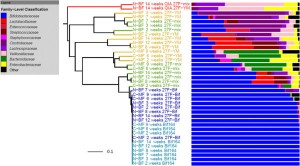
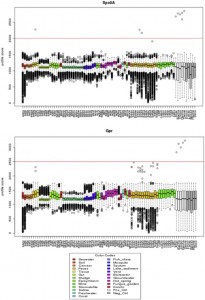
Another paper on how sample processing (and in this case PCR primer choice) can influence microbiome studies. And another one that is definitely worth looking at: 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 2015, 3:26 doi:10.1186/s40168-015-0087-4 by Alan W. Walker, Jennifer C. Martin, …
A moderately new paper is out that is an excellent example of how biases in DNA extraction can have major impacts on inferences from culture independent DNA studies. The paper is in what I think is a generally non open access journal (Computational and Structural Biotechnology Journal) for fortunately has been made open access Source: …
Three weeks ago I stood in front of the 60 attendees of the STAMPS course and asked, “How many of you are currently working with shotgun metagenomes?” Ten to fifteen people raised their hands. In contrast, almost all had their hands in the air when I asked how many were expecting to work with shotgun …
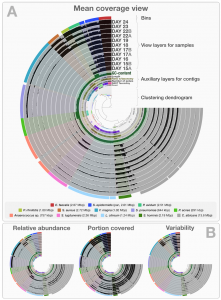
There is an interesting paper out a few days ago in PeerJ: MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. By Dongwan D. Kang, Jeff Froula, Rob Egan, Zhong Wang​. The key to what they do in the paper is summarized in Figure 1: The legend is below: There are three preprocessing steps before MetaBAT is …
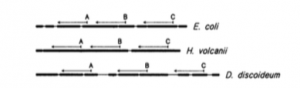
I got this email a few days ago from Jed Fuhrman. He had sent it to a group of people working on mcirobial diversity and he encouraged people to share it. I asked and he approved posting it here and I thought it would be of interest. (The image above is from Lane et al. 1985 …
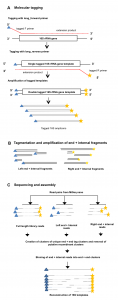
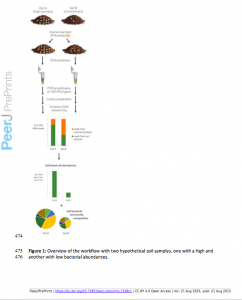
A new preprint has come out in PeerJ that presents a simple yet elegant solution to one of the bigger problems in DNA based microbial diversity studies. The authors (Wenke Smets, Jonathan W Leff, Mark A Bradford, Rebecca L McCulley, Sarah Lebeer, Noah Fierer) show how one can use the addition of an internal standard …
So – I keep getting asked about this. I am looking for how people are doing DNA extractions from 1000s of samples. Some questions 1. What are the cheapest service providers out there for doing DNA extractions from 1000s of samples? 2. What would be the best way to do this oneself? That is, if you were …
About a year ago, I posted here about my free, interactive text: An Introduction to Applied Bioinformatics. The project is now Sloan-funded (as of April 2015), so there will be a lot of changes and expanded content coming down the pipe over the next few months. One major change that went in recently, and which …