Blog post prepared jointly by Andrew Doxey (@acdoxey) and Josh Neufeld (@joshdneufeld) The “aquariome” Back in 2013, as part of a project assessing aquarium microbial communities and their role in nutrient cycling, Laura Sauder (graduate student in the Neufeld lab) sequenced a shotgun metagenomic library from a freshwater aquarium biofilter that was installed on this …
Here is a new set of papers that came out in the past week(s) that I posted at MicrobiomeDigest, but that I also wanted to share here. Three of these papers are from BMC’s Microbiome journal, which recently has published several other built environment microbiology papers, so it’s worth checking out. Microbes in buildings Moisture …
When we think of the built environment, we usually only include the indoor surfaces and air. A new study (preprint) posted last week on BioRxiv went through the roof, and looked at the microbes living on solar panels. A highly diverse, desert-like microbial biocenosis on solar panels in a Mediterranean city – Pedro Dorado-Morales, Cristina Vilanova, Juli …
Norm Pace gave a talk at UC Davis yesterday on “Metagenomics and the Tree of Life”. I and a few other people posted live Tweets from the talk which I have compiled together via the Storify system. This “Storify” is embedded below. In addition, Lisa Cohen, who was at the talk posted her notes which …
Special Seminar Dr. Norman Pace Distinguished Professor Department of Molecular, Cellular and Developmental Biology University of Colorado, Boulder Metagenomics and the Tree of Life Tuesday, October 6th, 2015 1.30 pm Genome and Biomedical Science Facility (GBSF) 1005 Host: Jonathan Eisen (jaeisen@ucdavis.edu) For more about Pace’s work see http://pacelab.colorado.edu
Some antibiotics routinely used in the clinic are considered to have little side effects. Such is the case for cefprozil, a second generation cephalosporin. This relative safety may be true (to a certain degree) from the viewpoint of humans taking the antibiotic. However, it is not the case from our commensal microbes’ perspective. Our study, …
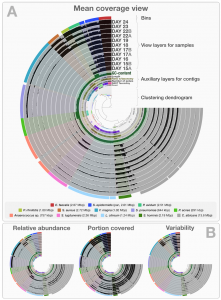
Three weeks ago I stood in front of the 60 attendees of the STAMPS course and asked, “How many of you are currently working with shotgun metagenomes?” Ten to fifteen people raised their hands. In contrast, almost all had their hands in the air when I asked how many were expecting to work with shotgun …
I just saw this paper, published a couple of days ago in Nature’s Scientific Reports. And yeah, it’s open access! While reading this post, I would suggest playing Dalai Lama by Rammstein, the in-flight version of Der Erlkönig. Meta-genomic analysis of toilet waste from long distance flights; a step towards global surveillance of infectious diseases and antimicrobial …
So … what goes around comes around. In 2003 and 2004, I spent a lot of time discussing and arguing with people about what would be the best strategy for making and sequencing Sanger libraries for metagenomic sequencing for the Sargasso Sea metagenome study coordinated by the Venter Institute (I worked at TIGR at the time and …
There is an interesting and potentially very useful paper just out: Optimized DNA extraction and metagenomic sequencing of airborne microbial communities A brief summary from the journal: This protocol enables collection of airborne particulate matter; and after sample pretreatment, it allows sufficient quantities of microbial DNA to be extracted and prepared for downstream applications such …