My collaborator, Marina LaForgia, and I are gearing up to start processing some plant rhizosphere samples for metagenomic sequencing. We collected more samples than we can afford to process (as is life) and are starting to make the hard decisions about which samples to move forward with. This brought us to the topic of whether …
New report on Synthetic Biology from the National Academies of Sciences, Engineering and Medicine that may be of interest. Synthetic biology expands the possibilities for creating new weapons — including making existing bacteria and viruses more harmful — while decreasing the time required to engineer such organisms, concludes a new report by the National Academies …
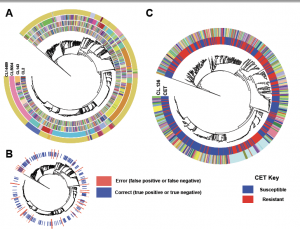
This preprint seems like it may be of interest to folks: Precise prediction of antibiotic resistance in Escherichia coli from full genome sequences | bioRxiv Basically, the authors showed that, using a machine learning approach, they can quite accurately predict antibiotic resistance in E. coli from whole genome data. The emergence of microbial antibiotic resistance …
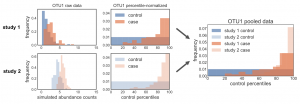
We recently published a method to normalize data that corrects for batch effects and allows for pooling of raw data across studies. In this post, I want to discuss more about the important (but potentially confusing) aspect of our method, where we add noise to the zeros before performing the normalization. I’ve gotten a few …
Twitter Logo: Wikimedia Commons Public Domain Thought this might be of interest to various folks. I made a Twitter moment regarding a discussion about microbial genome annotation methods. Microbial Genome Annotation Methods https://platform.twitter.com/widgets.js
The Metcalf lab at Colorado State University is seeking a microbiome bioinformatics postdoctoral scientist. The scientist will analyze multi-omics data sets, including but not limited to: amplicon sequencing data, shotgun metagenomics data, and metabolomics data. Min. reqs. include: Ph.D. in bioinformatics, ecology, microbiology or similar field. Proficient in coding languages (Python preferred). Demonstrated knowledge of …
So – just a quick post for now. More details to come. But I wanted to get a little bit about this out there. For a few years I have been teaching a mid-level course at UC Davis on “DNA sequencing based studies of microbial diversity’. The course has evolved over the years from a …
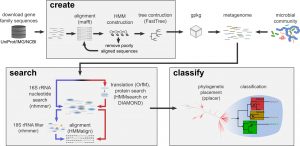
This could be of use to many people: Source: GraftM: a tool for scalable, phylogenetically informed classification of genes within metagenomes | Nucleic Acids Research | Oxford Academic Large-scale metagenomic datasets enable the recovery of hundreds of population genomes from environmental samples. However, these genomes do not typically represent the full diversity of complex microbial …
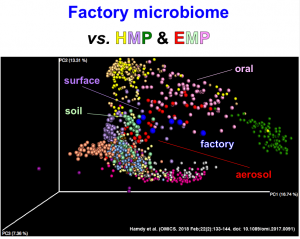
The story behind: Toward the Drug Factory Microbiome: Microbial Community Variations in Antibiotic-Producing Clean Rooms (OMICS. 2018 Feb;22(2):133-144. doi: 10.1089/omi.2017.0091.) PMID: 28873001 [Preprint] It was in 2013, on my return to Cairo University after a couple of years as a visiting scientist at UCSD, that I met Amal for the first time. Amal was a fresh master’s …
(Note: This post was updated on 3/26/18 after re-running some of our analysis based on suggestions in the comments and on Twitter). We were very excited to get our MinION sequencer from Nanopore 1.5 years ago… so many potential applications; sequencing in real time, use in the classroom, use in the field, not having to …